Volume 24, Number 10—October 2018
Research
Molecular Evolution, Diversity, and Adaptation of Influenza A(H7N9) Viruses in China
Figure 5
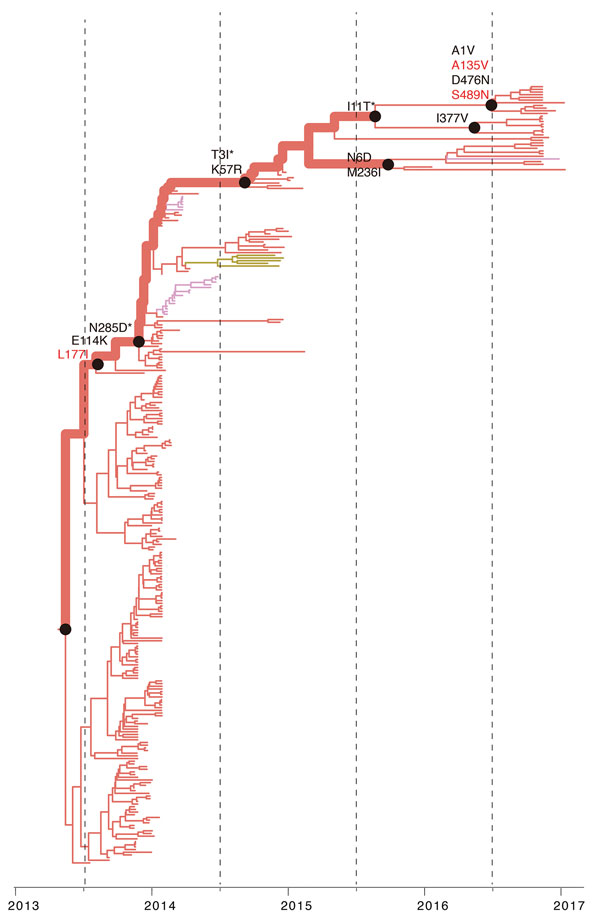
Figure 5. Reconstruction of amino acid changes along trunk of lineage C phylogenies of influenza A(H7N9) viruses, China. Maximum clade credibility tree of hemagglutinin gene sequences from lineage C is shown. Branches are colored according to geographic locations, as in Figure 3. Thicker lines indicate the trunk lineage leading up to the current fifth influenza epidemic wave. Amino acid changes along the trunk are indicated. Red branches indicate sites undergoing parallel amino acid changes across multiple lineages. Mutations correspond to H3 numbering scheme. *Mutation sites not present are numbered according to H7 numbering.
1These authors contributed equally to this article.
2These senior authors jointly supervised this study.
Page created: September 14, 2018
Page updated: September 14, 2018
Page reviewed: September 14, 2018
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.