Volume 24, Number 3—March 2018
Research
Capsule Typing of Haemophilus influenzae by Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry1
Abstract
Encapsulated Haemophilus influenzae strains belong to type-specific genetic lineages. Reliable capsule typing requires PCR, but a more efficient method would be useful. We evaluated capsule typing by using matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry. Isolates of all capsule types (a−f and nontypeable; n = 258) and isogenic capsule transformants (types a−d) were investigated. Principal component and biomarker analyses of mass spectra showed clustering, and mass peaks correlated with capsule type-specific genetic lineages. We used 31 selected isolates to construct a capsule typing database. Validation with the remaining isolates (n = 227) showed 100% sensitivity and 92.2% specificity for encapsulated strains (a−f; n = 61). Blinded validation of a supplemented database (n = 50) using clinical isolates (n = 126) showed 100% sensitivity and 100% specificity for encapsulated strains (b, e, and f; n = 28). MALDI-TOF mass spectrometry is an accurate method for capsule typing of H. influenzae.
Haemophilus influenzae is subdivided into encapsulated strains, which express different serotypes of capsular polysaccharide (designated types a–f), and nonencapsulated strains, which are designated nontypeable H. influenzae (NTHi) (1). Since the introduction of conjugate vaccines against H. influenzae type b (Hib), a common cause of meningitis, epiglottitis, and sepsis in small children, the epidemiology of invasive H. influenzae disease has changed dramatically, with an increase in the diversity of serotypes responsible for illness.
Although the incidence of Hib disease has decreased in countries implementing childhood vaccination (2), invasive disease caused by NTHi has become more prominent during the same period, especially among newborns and the elderly (3–6). In the postvaccination era, increasing incidences and outbreaks of invasive H. influenzae type a (Hia) infections have been reported in South and North America (7–10), particularly among the indigenous populations in Canada and the United States (7,8,10). Studies have also suggested an increase in cases of invasive H. influenzae type e (Hie) and type f (Hif) disease (4,11,12). Hib vaccine failures have been described (13), and omission of booster dose(s) appears to result in a rapidly increased incidence of invasive Hib disease (14,15), suggesting continued circulation of Hib isolates in the community. Globally, one third of eligible children still do not receive adequate vaccination (16).
Encapsulated H. influenzae strains are generally genetically clonal. This finding was first demonstrated by multilocus enzyme electrophoresis, by which encapsulated isolates could be separated into different genetic lineages that correspond to different capsule types (17). This clonal population structure has been confirmed by multilocus sequence typing (MLST), which assigns isolates to different sequence types (STs), although some differences have been observed in the organization of different lineages (18). There are 3 known major genetic groups of Hia and Hib (18,19). Two genetic groups of Hia (related to ST21 and ST23) account for most Hia isolates in the MLST database (20). For Hib, ST6-related isolates account for most cases (17,18), whereas the second most common genetic group is related to ST222 (18,21). There is 1 known lineage each for serotypes c through f (18,19). In contrast, NTHi are genetically heterogenous (19).
Capsule typing of H. influenzae has traditionally been performed by using slide agglutination with antisera (conventional serotyping), but incorrect results are common, and specificity for encapsulated isolates is low (22,23). Determination of presence of the capsule gene complex (bexA or bexB) by PCR, followed by type-specific cap a–f PCRs has excellent sensitivity and specificity but is laborious and time-consuming (24–27). Because of limitations of current typing methods, typing might be delayed or not performed in clinical practice. However, rapidly obtained information on capsule type is still of interest for the treating clinician (5) and, in particular, for monitoring of capsule type distribution and effectiveness of Hib vaccination programs, especially with respect to invasive disease.
Matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry is commonly used to identify bacterial and fungal species, including H. influenzae, by analyzing the composition of ribosomal proteins in a sample. It is a rapid and convenient method and has a low cost per sample (28). Recently, we have shown that MALDI-TOF mass spectrometry can separate Hib from non-b H. influenzae (29). In this study, we examined the capacity of MALDI-TOF mass spectrometry to perform full capsule typing of H. influenzae. This method would be valuable for first-line diagnostics of H. influenzae to identify patients at risk for immunodeficiency or anatomic cerebrospinal fluid space defect, and to detect rapidly outbreaks caused by specific capsule types. It would also increase time and cost effectiveness of surveillance of H. influenzae epidemiology and Hib vaccination efficacy.
Bacterial Isolates
We used 2 culture collections in this study (Figure 1). The first collection was an evaluation set of isolates used to construct a coherent reference database and was composed of 258 H. influenzae strains. It included isolates from 3 major clinical laboratories in Sweden (Malmö/Lund, Gothenburg, and Stockholm) obtained in 1997–2011 but also a wide range of international strains from different countries, continents, and time periods (n = 41; Technical Appendix Table). In addition, we included 4 isogenic capsule-transformed strains of types a (Rb–/a+:02), b (Rb+:02), c (Rb–/c+:02), and d (Rb–/d+:02) (30) in the study. These strains originate from strain Rd, a capsule-deficient type d strain (31). For validation of the new MALDI-TOF mass spectrometry typing method, we used a second collection composed of 126 bloodstream and cerebrospinal fluid H. influenzae isolates obtained in Sweden during 2010 and 2013–2016. All isolates were identified as H. influenzae by using standard laboratory taxonomy techniques and were grown on chocolate agar plates overnight (18–24 h) in a humid atmosphere at 37°C containing 5% CO2 before any experiments were conducted.
PCR for Capsule Typing and MLST
We prepared DNA by adding a few colonies of bacteria to distilled water. After heating at 98°C for 10 min, we centrifuged each sample at 16,000 × g for 5–10 min and collected the supernatant. In a few instances, we extracted DNA by using the GenElute Bacterial Genomic DNA Kit (Sigma-Aldrich, St. Louis, MO, USA) according to the manufacturer’s instructions. We performed capsule typing by PCR using bexB and type-specific cap primers for all isolates as described (24–26).
We performed PCR for MLST genes as described (18) and sequenced the resulting PCR products by using the forward primer and, if necessary for adequate sequence quality, the reverse primer. We trimmed and edited sequences before concatenation (total length 3,057 bp). We deposited MLST nucleotide sequences in GenBank (accession nos. MG550316–MG550889). Some isolates had been previously typed by MLST. In these instances, we retrieved MLST data from the MLST database (20).
Analysis of MLST Data
We determined sequence types by using the MLST database. We aligned concatenated sequences in Geneious 9.1.8 (Biomatters, Auckland, New Zealand) and used the PAUP* 4.0a158 plug-in (http://phylosolutions.com/paup-test/) to construct a maximum-likelihood phylogenetic tree. The best fitting model was estimated to be the generalized time-reversible model including invariant sites and gamma distribution by using the Akaike information criterion in jModelTest 2.1.10 (32,33). We visualized the resulting tree by using FigTree 1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/). All isolate and ST information has been submitted to the MLST database.
Acquisition of MALDI-TOF Mass Spectrometry Data
We acquired mass spectra by using a Microflex LT MALDI-TOF mass spectrometry system (Bruker Daltonics, Bremen, Germany), with default settings as described (29). We prepared all isolates for acquisition of spectra by using the ethanol–formic acid procedure described by the instrument manufacturer. We spotted isolates on 2 spots and analyzed each spot 3 times, resulting in 6 spectra/isolate. Isolates in the reference database were spotted on 8 spots, resulting in 24 spectra, before being added to the database.
Analysis of MALDI-TOF Mass Spectrometry Data
In Mass-Up 1.0.13 (34), we preprocessed and analyzed raw spectra of all isolates in the evaluation set (n = 258) and capsule transformants (n = 4) by using the integrated MALDIquant analysis package for R (http://strimmerlab.org/software/maldiquant/). We performed preprocessing with intensity transformation (square root), smoothing (Savitzky–Golay), baseline correction (Top-Hat), and intensity standardization (total ion current). We performed peak detection with a signal-to-noise ratio of 2, a half window size of 50, and no minimum peak intensity. We calculated a consensus spectrum for each isolate with a peak tolerance of 0.002 and percentage of presence of 60%. For principal component analysis (PCA) (35) and biomarker analysis, we performed intersample matching with a peak tolerance of 0.002. PCA was performed with default settings (maximum number of components = −1 and 0.95 of the total variance covered). In the biomarker analysis, we calculated a p value for each peak by using the randomization test of independence.
Construction and Validation of a MALDI Biotyper Database for Capsule Typing
We used selected isolates from the evaluation set to create main spectra (MSPs) for a new MALDI Biotyper 4.1 database (Bruker Daltonics) (Figure 1). These reference isolates were selected to represent all capsule types and genetic lineages. NTHi strains were selected with the aim of including isolates of all known genetic clades (19). Spectra of reference isolates were controlled by using FlexAnalysis (Bruker Daltonics). We performed smoothing (Savitzky-Golay) and baseline correction (Top-Hat) and excluded spectra with outlier appearance (lacking or having an extra peak) and low quality (peaks outside a 500 ppm range). If <20 spectra remained after the control, new spectra for that specific isolate were obtained. We used default settings for spectra preprocessing, MSP creation, and identification as described (29).
We used isolates in the evaluation set not selected as reference isolates for initial validation of the database (Figure 1). Because all isolates were H. influenzae, high score values (>2.0) were expected. Thus, we classified each spectrum according to the top matching MSP in the new database. For isolate classification, >5/6 spectra classified to the same type (a–f or NTHi) were required. If <4/6 spectra were classified to the same type, the isolate was classified as inconclusive. To improve the specificity of the typing method, considering the known heterogeneity of NTHi, we supplemented the capsule typing database with NTHi isolates not correctly classified in the initial validation until all isolates in the evaluation set not included in the database were correctly classified on every single spectrum. Finally, we blindly validated the supplemented database by using MALDI-TOF mass spectrometry classification of invasive isolates (n = 126) obtained during 2010 and 2013–2016 and calculated sensitivity and specificity by using PCR typing as the standard (Figure 1).
Genetic Lineages of Encapsulated H. influenzae in the Evaluation Set
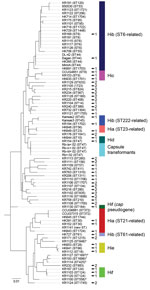
To construct a clinically useful reference database for capsule typing by MALDI-TOF mass spectrometry, we aimed to identify and collect isolates from all known lineages of encapsulated H. influenzae (Figure 2; online Technical Appendix Table). We performed MLST for all Hia (n = 9) and Hib isolates (n = 24) in the evaluation set, in addition to a subset of isolates of other capsule types (c–f), capsule transformants (n = 4), and NTHi. Phylogenetic analysis confirmed that the collection contained isolates from different genetic lineages of encapsulated H. influenzae, including the 2 major genetic groups of type a and all 3 lineages of type b (Figure 2). Capsule transformants belonged to the known genetic lineage of Hid isolates (Figure 2) and were the same ST as the parental strain Rd (18). One isolate (KR1130) was typed by PCR as Hif (bexB- and cap f-positive) but phylogenetically belonged to a lineage separate from all other Hif isolates. Thus, this isolate was not part of the established, ST124-related Hif lineage (Figure 2). The cap locus of this isolate was sequenced and found to be a nonexpressed pseudogene (data not shown).
MALDI-TOF Mass Spectrometry of Genetic Lineages of Encapsulated H. influenzae
We performed PCA for all isolates in the evaluation set (n = 258) and the capsule transformants (n = 4). As expected, NTHi formed a large heterogeneous group, but clustering of encapsulated isolates of the same capsule types was found (Figure 3, panel A). When PCA was performed on encapsulated isolates (n = 83) and capsule transformants (n = 4) only, the clustering became clearer and was particularly evident for Hib, Hie, and Hif isolates (Figure 3, panel B). Encapsulated isolates segregated in groups according to capsule type and, for Hia and Hib isolates, by genetic lineage according to MLST (Figure 2; 3, panel B). Capsule transformants were found as a separate group in close proximity of Hid isolates and not distributed according to their respective capsule type (Figure 3, panel B). Isolate KR1130 did not cluster with Hif isolates of the ST124-related lineage (Figure 3, panel B).
Biomarker analysis of encapsulated isolates and capsule transformants identified several peaks conserved within the different genetic lineages of capsule types, indicating the possibility of separating them on the basis of MALDI-TOF mass spectra (Figure 4). Capsule transformants expressed similar peak patterns relative to each other but differed in many peaks when compared with wild-type strains of the same capsule types.
Sensitivity and Specificity of Automated Capsule Typing by MALDI-TOF Mass Spectrometry
An initial capsule typing reference database was constructed in MALDI Biotyper. Encapsulated isolates (n = 22) representing all major genetic lineages of encapsulated H. influenzae were included (Figure 2). To ensure adequate coverage of potential variation within each lineage, multiple reference isolates were chosen for each lineage (when possible) on the basis of geographic origin and variations in mass spectra. In addition, NTHi (n = 9) representing 8 of 10 known genetic clades of NTHi (19) were included in the database (Figure 2).
Validation of the original database (n = 31) using the remaining isolates in the evaluation set (n = 227) showed 100% sensitivity for encapsulated isolates (Table 1), and every isolate was correctly classified on every spectrum. All capsule transformants were classified as type d, the original serotype of the parental strain Rd (31). No isolate matched KR1130, the isolate typed by PCR as Hif with a pseudogene cap locus.
A few NTHi were either inconclusively typed or misclassified as encapsulated, resulting in reduced specificity for encapsulated isolates (Table 1). For this reason, we supplemented the capsule typing database with misclassified NTHi from the evaluation set until the database correctly classified all the remaining isolates in the evaluation set on every single spectrum. This modification resulted in an additional 19 NTHi being added to the MALDI Biotyper database (Figure 2). When PCA was performed separately for NTHi in the evaluation set, it was evident that the supplemented database covered the heterogeneity of NTHi better than the original database (Figure 5). The same finding was evident from phylogenetic analysis (Figure 2). Five NTHi were misclassified as Hie and could not be added to the database because they interfered with classification of true Hie isolates and would decrease sensitivity for Hie (Figure 2).
As a final performance test, we blindly validated the supplemented database (n = 50) by using a separate culture collection consisting of clinical invasive isolates from Sweden (n = 126) obtained during 2010 and 2013–2016. When we compared MALDI-TOF mass spectrometry capsule typing results with PCR capsule typing results, all encapsulated isolates (types b, e, and f; n = 28) were correctly classified on every single spectrum (Table 2). Of 98 NTHi, only 5 were not correctly classified. These isolates were all classified as inconclusive. Thus, no NTHi was incorrectly classified as encapsulated, and the resulting sensitivity and specificity of capsule typing was 100% in the final validation (Table 2).
In this study, we have shown that encapsulated H. influenzae have different MALDI-TOF mass spectra that correlate with genetic lineages representing different capsule types. We have demonstrated that, after construction of a comprehensive reference database, routine MALDI-TOF mass spectrometry analysis has excellent capacity for identifying type-specific genetic lineages associated with encapsulated H. influenzae and thereby can be used for capsule typing of H. influenzae.
Our study had several strengths. We analyzed a large collection of well-characterized strains collected at different times from various geographic regions to ensure the robustness of our findings. Using MLST, we ensured adequate coverage of the major genetic lineages of encapsulated H. influenzae in the MALDI-TOF mass spectrometry reference database. Moreover, the database was carefully evaluated and supplemented to ensure adequate coverage of the heterogeneity of NTHi. We blindly validated the supplemented database to mimic an authentic clinical or epidemiologic situation and demonstrated excellent sensitivity and specificity compared with conventional PCR-based typing. During construction of the capsule typing database, we identified several isolates previously typed by PCR or agglutination (by us or others) in which the MALDI-TOF mass spectrometry results did not match the suggested capsule type. When we retyped these isolates by PCR, the capsule type suggested by MALDI-TOF mass spectrometry proved to be correct in all instances (except for the NTHi typed as Hie) (Figure 2), and isolates were reassigned to a new capsule type, further supporting the capacity of MALDI-TOF mass spectrometry for capsule typing.
Our study had some limitations. The first limitation reflects the limited availability of some rare variants. Our collection contained no Hia isolates belonging to the uncommon ST4-related genetic group. For the ST61-related lineage of Hib, we had access to only 1 isolate, which was included in the reference database and thus not represented in the test collection. However, our ST61 isolate was separable when mass spectra were analyzed by PCA and biomarker analysis, and no isolate was misclassified to this lineage in the initial or final validation of the typing databases. Furthermore, we have demonstrated that identification of the ST222-related Hib lineage by MALDI-TOF mass spectrometry is possible (Table 1), which was not the case previously (29). The second potential limitation arises through the genetic heterogeneity of NTHi, making adequate representation in the reference database a challenge (19). This limitation was apparent during the initial evaluation of the typing method, when some NTHi were misclassified. To address this issue, we supplemented the database with 19 additional reference NTHi strains. The final validation of our typing method demonstrated excellent specificity for NTHi, but the sensitivity for identifying encapsulated isolates remained unchanged. Because most invasive infections in countries implementing Hib vaccination are caused by NTHi, a high specificity is desirable (3–5).
MALDI-TOF mass spectrometry has proved valuable in subtyping several clinically relevant bacteria, including Clostridium difficile (36), methicillin-resistant Staphylococcus aureus (37,38), and enterohemorrhagic Escherichia coli (39). Subtyping generally relies on common genetic differences between isolates, reflected in the composition of the proteins measured. In our study, wild-type isolates of different capsule types could be separated, but isogenic capsule transformants could not. These isolates were classified as type d, the original capsule type of the parental strain Rd. This finding confirms that capsule type identification is based on a proxy identification of genetic lineage, rather than identification of capsule biosynthesis-associated proteins. Thus, our method is an indirect typing method, as opposed to serotyping, which identifies the capsule polysaccharide, and PCR, which identifies the capsule gene complex directly.
Although there is little evidence that new lineages of encapsulated H. influenzae have appeared historically, novel lineages of encapsulated strains might appear and be missed by the method. Isolate KR1130 used in this study was initially suspected to represent such a lineage. However, its cap locus was shown to be on a nonexpressed pseudogene. Only 1 other isolate of the same ST (ST184) is currently registered in the MLST database, and it is a nontypeable isolate. No other Hif strain in this study or the MLST database belongs to this genetic lineage (20).
One advantage of indirect capsule type identification by MALDI-TOF mass spectrometry is that determination of genetic lineage of encapsulated isolates can be made without further analysis. The method can also identify previously encapsulated capsule-deficient strains, which have lost parts or all of the cap locus, either during infection or laboratory handling (40–42).
A concern regarding subtyping by MALDI-TOF mass spectrometry (43) is the potential need for special sample preparations, such as growth conditions and type of matrix. In several studies, differences in mass spectra between subtypes of various species were observed but no automated classification methods were reported (43), which might limit general applicability. In this study, we used standard growth conditions, as well as routine ethanol–formic acid extraction and mass spectra acquisition protocols. The software used (MALDI Biotyper) also has the advantage of being a standard software used in clinical settings. These factors greatly increased the chance of clinical implementation of our findings.
In conclusion, our study demonstrated that rapid capsule typing of H. influenzae by identification of capsule type-specific genetic lineages using routine MALDI-TOF mass spectrometry is possible and highly accurate. After further large-scale validation, this method has the potential for clinical and research use. With the increasing heterogeneity in capsule types of disease-causing H. influenzae observed since Hib conjugate vaccines were introduced, the method can become a valuable tool in clinical diagnostic laboratories.
Dr. Månsson is a physician and doctoral student in clinical microbiology at the Riesbeck Laboratory, Lund University, Malmö, Sweden. His main research interests are respiratory tract infections, with special focus on H. influenzae diagnostics, epidemiology, and antimicrobial drug resistance.
Acknowledgment
This study was supported by grants from the Foundations of Anna and Edwin Berger (to K.R.); the Royal Physiographical Society of Lund, (Forssmans Foundation (to V.M.); the Swedish Medical Research Council (grant no. K2015-57X-03163-43-4; http://www.vr.se); and the Skåne County Council Research and Development Foundation (to K.R.).
References
- Pittman M. Variation and type specificity in the bacterial species Haemophilus influenzae. J Exp Med. 1931;53:471–92. DOIPubMedGoogle Scholar
- Peltola H. Worldwide Haemophilus influenzae type b disease at the beginning of the 21st century: global analysis of the disease burden 25 years after the use of the polysaccharide vaccine and a decade after the advent of conjugates. Clin Microbiol Rev. 2000;13:302–17. DOIPubMedGoogle Scholar
- Langereis JD, de Jonge MI. Invasive disease caused by nontypeable Haemophilus influenzae. Emerg Infect Dis. 2015;21:1711–8. DOIPubMedGoogle Scholar
- Resman F, Ristovski M, Ahl J, Forsgren A, Gilsdorf JR, Jasir A, et al. Invasive disease caused by Haemophilus influenzae in Sweden 1997-2009; evidence of increasing incidence and clinical burden of non-type b strains. Clin Microbiol Infect. 2011;17:1638–45. DOIPubMedGoogle Scholar
- Ladhani S, Slack MP, Heath PT, von Gottberg A, Chandra M, Ramsay ME; European Union Invasive Bacterial Infection Surveillance participants. Invasive Haemophilus influenzae Disease, Europe, 1996-2006. Emerg Infect Dis. 2010;16:455–63. DOIPubMedGoogle Scholar
- Whittaker R, Economopoulou A, Dias JG, Bancroft E, Ramliden M, Celentano LP; European Centre for Disease Prevention and Control Country Experts for Invasive Haemophilus influenzae Disease. Epidemiology of Invasive Haemophilus influenzae Disease, Europe, 2007-2014. Emerg Infect Dis. 2017;23:396–404. DOIPubMedGoogle Scholar
- Bender JM, Cox CM, Mottice S, She RC, Korgenski K, Daly JA, et al. Invasive Haemophilus influenzae disease in Utah children: an 11-year population-based study in the era of conjugate vaccine. Clin Infect Dis. 2010;50:e41–6. DOIPubMedGoogle Scholar
- Bruce MG, Zulz T, DeByle C, Singleton R, Hurlburt D, Bruden D, et al. Haemophilus influenzae serotype a invasive disease, Alaska, USA, 1983-2011. Emerg Infect Dis. 2013;19:932–7. DOIPubMedGoogle Scholar
- Zanella RC, Bokermann S, Andrade AL, Flannery B, Brandileone MC. Changes in serotype distribution of Haemophilus influenzae meningitis isolates identified through laboratory-based surveillance following routine childhood vaccination against H. influenzae type b in Brazil. Vaccine. 2011;29:8937–42. DOIPubMedGoogle Scholar
- Rotondo JL, Sherrard L, Helferty M, Tsang R, Desai S. The epidemiology of invasive disease due to Haemophilus influenzae serotype a in the Canadian North from 2000 to 2010. Int J Circumpolar Health. 2013;72:72. DOIPubMedGoogle Scholar
- Bajanca-Lavado MP, Simões AS, Betencourt CR, Sá-Leão R; Portuguese Group for Study of Haemophilus influenzae invasive infection. Characteristics of Haemophilus influenzae invasive isolates from Portugal following routine childhood vaccination against H. influenzae serotype b (2002-2010). Eur J Clin Microbiol Infect Dis. 2014;33:603–10. DOIPubMedGoogle Scholar
- Ladhani SN, Collins S, Vickers A, Litt DJ, Crawford C, Ramsay ME, et al. Invasive Haemophilus influenzae serotype e and f disease, England and Wales. Emerg Infect Dis. 2012;18:725–32. DOIPubMedGoogle Scholar
- Ladhani S, Heath PT, Slack MP, McIntyre PB, Diez-Domingo J, Campos J, et al.; Participants of the European Union Invasive Bacterial Infections Surveillance Network. Haemophilus influenzae serotype b conjugate vaccine failure in twelve countries with established national childhood immunization programmes. Clin Microbiol Infect. 2010;16:948–54. DOIPubMedGoogle Scholar
- Ladhani SN. Two decades of experience with the Haemophilus influenzae serotype b conjugate vaccine in the United Kingdom. Clin Ther. 2012;34:385–99. DOIPubMedGoogle Scholar
- Juarez MD, Rancaño C, Neyro S, Biscayart C, Katz N, Pasinovich M, et al. What’s happening with Haemophilus influenzae type B invasive disease in Latin America region? Argentina’s experience. In: Abstracts of IDWeek 2016, New Orleans, October 26–30. Abstract no. 768 [cited 2017 Dec 18]. https://idsa.confex.com/idsa/2016/webprogram/POSTER.html
- World Health Organization. Global and regional immunization profile, 2016 global summary [cited 2017 Nov 9]. http://www.who.int/immunization/monitoring_surveillance/data/gs_gloprofile.pdf?ua=1
- Musser JM, Kroll JS, Moxon ER, Selander RK. Clonal population structure of encapsulated Haemophilus influenzae. Infect Immun. 1988;56:1837–45.PubMedGoogle Scholar
- Meats E, Feil EJ, Stringer S, Cody AJ, Goldstein R, Kroll JS, et al. Characterization of encapsulated and noncapsulated Haemophilus influenzae and determination of phylogenetic relationships by multilocus sequence typing. J Clin Microbiol. 2003;41:1623–36. DOIPubMedGoogle Scholar
- Erwin AL, Sandstedt SA, Bonthuis PJ, Geelhood JL, Nelson KL, Unrath WC, et al. Analysis of genetic relatedness of Haemophilus influenzae isolates by multilocus sequence typing. J Bacteriol. 2008;190:1473–83. DOIPubMedGoogle Scholar
- University of Oxford. Haemophilus influenzae MLST website [cited 2017 Nov 9]. http://pubmlst.org/hinfluenzae/
- Myers AL, Jackson MA, Zhang L, Swanson DS, Gilsdorf JR. Haemophilus influenzae type b invasive disease in Amish children, Missouri, USA, 2014. Emerg Infect Dis. 2017;23:112–4. DOIPubMedGoogle Scholar
- LaClaire LL, Tondella ML, Beall DS, Noble CA, Raghunathan PL, Rosenstein NE, et al.; Active Bacterial Core Surveillance Team Members. Identification of Haemophilus influenzae serotypes by standard slide agglutination serotyping and PCR-based capsule typing. J Clin Microbiol. 2003;41:393–6. DOIPubMedGoogle Scholar
- Satola SW, Collins JT, Napier R, Farley MM. Capsule gene analysis of invasive Haemophilus influenzae: accuracy of serotyping and prevalence of IS1016 among nontypeable isolates. J Clin Microbiol. 2007;45:3230–8. DOIPubMedGoogle Scholar
- Falla TJ, Crook DW, Brophy LN, Maskell D, Kroll JS, Moxon ER. PCR for capsular typing of Haemophilus influenzae. J Clin Microbiol. 1994;32:2382–6.PubMedGoogle Scholar
- Davis GS, Sandstedt SA, Patel M, Marrs CF, Gilsdorf JR. Use of bexB to detect the capsule locus in Haemophilus influenzae. J Clin Microbiol. 2011;49:2594–601. DOIPubMedGoogle Scholar
- Lâm TT, Elias J, Frosch M, Vogel U, Claus H. New diagnostic PCR for Haemophilus influenzae serotype e based on the cap locus of strain ATCC 8142. Int J Med Microbiol. 2011;301:176–9. DOIPubMedGoogle Scholar
- van Ketel RJ, de Wever B, van Alphen L. Detection of Haemophilus influenzae in cerebrospinal fluids by polymerase chain reaction DNA amplification. J Med Microbiol. 1990;33:271–6. DOIPubMedGoogle Scholar
- Clark AE, Kaleta EJ, Arora A, Wolk DM. Matrix-assisted laser desorption ionization-time of flight mass spectrometry: a fundamental shift in the routine practice of clinical microbiology. Clin Microbiol Rev. 2013;26:547–603. DOIPubMedGoogle Scholar
- Månsson V, Resman F, Kostrzewa M, Nilson B, Riesbeck K. Identification of Haemophilus influenzae type b isolates by use of matrix-assisted laser desorption ionization−time of flight mass spectrometry. J Clin Microbiol. 2015;53:2215–24. DOIPubMedGoogle Scholar
- Zwahlen A, Kroll JS, Rubin LG, Moxon ER. The molecular basis of pathogenicity in Haemophilus influenzae: comparative virulence of genetically-related capsular transformants and correlation with changes at the capsulation locus cap. Microb Pathog. 1989;7:225–35. DOIPubMedGoogle Scholar
- Alexander HE, Leidy G. Determination of inherited traits of H. influenzae by desoxyribonucleic acid fractions isolated from type-specific cells. J Exp Med. 1951;93:345–59. DOIPubMedGoogle Scholar
- Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9:772. DOIPubMedGoogle Scholar
- Guindon S, Gascuel O, Rannala B. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. DOIPubMedGoogle Scholar
- López-Fernández H, Santos HM, Capelo JL, Fdez-Riverola F, Glez-Peña D, Reboiro-Jato M. Mass-Up: an all-in-one open software application for MALDI-TOF mass spectrometry knowledge discovery. BMC Bioinformatics. 2015;16:318. DOIPubMedGoogle Scholar
- Kehrmann J, Wessel S, Murali R, Hampel A, Bange FC, Buer J, et al. Principal component analysis of MALDI TOF MS mass spectra separates M. abscessus (sensu stricto) from M. massiliense isolates. BMC Microbiol. 2016;16:24. DOIPubMedGoogle Scholar
- Rizzardi K, Åkerlund T. High molecular weight typing with MALDI-TOF MS: a novel method for rapid typing of Clostridium difficile. PLoS One. 2015;10:e0122457. DOIPubMedGoogle Scholar
- Camoez M, Sierra JM, Dominguez MA, Ferrer-Navarro M, Vila J, Roca I. Automated categorization of methicillin-resistant Staphylococcus aureus clinical isolates into different clonal complexes by MALDI-TOF mass spectrometry. Clin Microbiol Infect. 2016;22:161.e1–7. DOIPubMedGoogle Scholar
- Josten M, Reif M, Szekat C, Al-Sabti N, Roemer T, Sparbier K, et al. Analysis of the matrix-assisted laser desorption ionization-time of flight mass spectrum of Staphylococcus aureus identifies mutations that allow differentiation of the main clonal lineages. J Clin Microbiol. 2013;51:1809–17. DOIPubMedGoogle Scholar
- Ojima-Kato T, Yamamoto N, Suzuki M, Fukunaga T, Tamura H. Discrimination of Escherichia coli O157, O26 and O111 from other serovars by MALDI-TOF MS based on the S10-GERMS method. PLoS One. 2014;9:e113458. DOIPubMedGoogle Scholar
- Fothergill LD, Chandler CA, Spencer M. Observations on the dissociation of meningitic strains of H. influenzae. J Immunol. 1936;31:401–15.
- Hoiseth SK, Connelly CJ, Moxon ER. Genetics of spontaneous, high-frequency loss of b capsule expression in Haemophilus influenzae. Infect Immun. 1985;49:389–95.PubMedGoogle Scholar
- Tsang RS, Li YA, Mullen A, Baikie M, Whyte K, Shuel M, et al. Laboratory characterization of invasive Haemophilus influenzae isolates from Nunavut, Canada, 2000-2012. Int J Circumpolar Health. 2016;75:29798. DOIPubMedGoogle Scholar
- Sauget M, Valot B, Bertrand X, Hocquet D. Can MALDI-TOF mass spectrometry reasonably type bacteria? Trends Microbiol. 2017;25:447–55. DOIPubMedGoogle Scholar
Figures
Tables
Cite This Article1Preliminary results from this study were presented at the IDWeek 2016 Conference, October 26−30, 2016, New Orleans, Louisiana, USA.
2These senior authors contributed equally to this article.
Table of Contents – Volume 24, Number 3—March 2018
| EID Search Options |
|---|
|
|
|
|
|
|




Please use the form below to submit correspondence to the authors or contact them at the following address:
Viktor Månsson, Riesbeck Laboratory, Department of Translational Medicine, Lund University, Jan Waldenströms gata 59, 205 02 Malmö, Sweden
Top