Volume 24, Number 3—March 2018
Research
Major Threat to Malaria Control Programs by Plasmodium falciparum Lacking Histidine-Rich Protein 2, Eritrea
Abstract
False-negative results for Plasmodium falciparum histidine-rich protein (HRP) 2–based rapid diagnostic tests (RDTs) are increasing in Eritrea. We investigated HRP gene 2/3 (pfhrp2/pfhrp3) status in 50 infected patients at 2 hospitals. We showed that 80.8% (21/26) of patients at Ghindae Hospital and 41.7% (10/24) at Massawa Hospital were infected with pfhrp2-negative parasites and 92.3% (24/26) of patients at Ghindae Hospital and 70.8% (17/24) at Massawa Hospital were infected with pfhrp3-negative parasites. Parasite densities between pfhrp2-positive and pfhrp2-negative patients were comparable. All pfhrp2-negative samples had no detectable HRP2/3 antigen and showed negative results for HRP2-based RDTs. pfhrp2-negative parasites were genetically less diverse and formed 2 clusters with no close relationships to parasites from Peru. These parasites probably emerged independently by selection in Eritrea. High prevalence of pfhrp2-negative parasites caused a high rate of false-negative results for RDTs. Determining prevalence of pfhrp2-negative parasites is urgently needed in neighboring countries to assist case management policies.
Eritrea, which is located in the Horn of Africa, has reduced malaria mortality and incidence rates extensively over the past decade (1,2). This reduction is attributed largely to integrated vector management, early diagnosis, and effective treatment implemented by the national malaria control program (NMCP). However, malaria remains a public health concern because there were ≈65,000 cases and 5 million persons living in malaria-prone areas in 2015 (3). Malaria is unstable in Eritrea, and seasonal and transmission patterns vary across 3 ecologic zones. Plasmodium falciparum infection accounts for ≈70% of confirmed malaria cases and P. vivax for the remaining ≈30% (4).
Microscopy remains the mainstay of malaria diagnosis at hospitals. Rapid diagnostic tests (RDTs) were introduced at the community level and primary health facilities in 2006, which paved the way for implementation of artemisinin-based combinations as first-line treatment in 2007. RDTs that detect P. falciparum and P. vivax simultaneously by targeting histidine-rich protein 2 (HRP2) and P. vivax plasmodium lactate dehydrogenase (pLDH), and met the World Health Organization (WHO)–recommended procurement criteria (5) were implemented in Eritrea with a quality assurance program that included training operators regularly and testing RDT lots before distribution.
In 2014, the NMCP received reports of false-negative RDT results in microscopically confirmed cases of P. falciparum and P. vivax malaria at several health facilities. The RDTs involved were from 10 lots that had passed testing at a WHO–Foundation for Innovative New Diagnostics lot-testing laboratory. To address this issue, the Ministry of Health (MOH) conducted exploratory investigations at 12 health facilities located in 4 regions of Eritrea and used different brands of RDTs. Results showed an overall false-negative result rate of 80% (41/50) for microscopically confirmed cases of P. falciparum malaria (6).
Deficiencies in RDT storage and operational issues were ruled out as causes of the false-negative results. Furthermore, samples of deployed RDTs were retrieved from the field and retested at a WHO–Foundation for Innovative New Diagnostics lot-testing laboratory against well-characterized reference samples. All RDTs passed testing, further suggesting that RDT quality issues did not cause false-negative results for P. falciparum (6). The cause of false-negative results for P. vivax was believed to be low parasite density. On the basis of these findings, the MOH recalled the RDTs and investigated P. falciparum–specific parasite factors, such as presence of P. falciparum lacking HRP2, as a primary cause.
HRP2-based RDTs target parasite HRP2, and some cross-react with HRP3 because of sequence similarities between the 2 proteins. P. falciparum lacking HRP2 and HRP3 caused by deletions of genes encoding these antigens (pfhrp2 and pfhrp3) in clinical cases were first reported in the Amazon region of Peru (7) and subsequently in Colombia (8), Suriname (9), and Brazil and Bolivia (10) (prevalence 4%–41%). The high prevalence of pfhrp2-negative parasites in Peru correlated with poor performance of HRP2-based RDTs (11) and led WHO to recommend using non–HRP2-based RDTs for case management in affected areas.
Outside South America, sporadic and low prevalence of pfhrp2-negative parasites has been reported in India (12); along the China–Myanmar border (13); and in countries in Africa, including Mali (14), Senegal (15), and the Democratic Republic of the Congo (16). A high prevalence (22%–41%) of pfhrp2-negative parasites was reported in 2 areas of Ghana (17). However, pfhrp2-negative parasites have not been detected in East Africa.
Unlike South America and Asia, where P. vivax infections dominate, most countries in Africa have predominantly P. falciparum infections. Thus, many of these countries have adopted HRP2-based RDTs only for P. falciparum. These RDTs do not detect pfhrp2-negative parasites, which results in P. falciparum infections not being detected and treated. Use of only HRP2-based RDTs in areas where pfhrp2-negative parasites are present will lead to increases in disease burden and transmission (18). Therefore, emergence of pfhrp2-negative parasites poses a serious threat to malaria control programs in Africa where malaria disease burden is high and RDT is the preferred diagnostic test. We investigated P. falciparum lacking pfhrp2 and pfhrp3 in Eritrea to determine whether pfhrp2/pfhrp3 deletions were a cause of false-negative results for HRP2-based RDTs and to describe origins of parasites.
Study Site
Eritrea is divided into 6 administrative regions. This study was conducted at Ghindae Hospital and Massawa Hospital in the Northern Red Sea Region (Figure 1) in March 2016. These locations were selected because of the timing of the malaria transmission season, easy access to infrastructure, and a high incidence of false-negative RDT results reported in this region (6).
Patient Enrollment and Sample Collection
Consecutive nonpregnant persons >5 years of age who came to the 2 hospitals and confirmed as being infected with P. falciparum by microscopy and a pan-pLDH single test line RDT (Carestart Malaria pLDH(PAN) G0111; Access Bio, Inc., Somerset, NJ, USA) were invited to participate. Venous blood (50 µL) was obtained from each consenting patient, spotted onto Whatman (Brentford, UK) 3M filter paper, dried, stored in a zip-lock bag containing desiccant, and used to prepare thin and thick blood smears. Smears and dried blood spots were shipped to different laboratories for further analyses.
Microscopy and HRP2-Based RDT
Blood smears were stained and examined at both hospitals and the Eritrean National Reference Laboratory (NRL; Asmara, Eritrea) according to the WHO microscopy manual (19). Final parasite species and density counts were determined by NRL microscopists who participated in the WHO/National Institute for Communicable Diseases malaria microscopy proficiency testing program. Blood samples were tested by using a combination HRP2/P. falciparum pLDH/P. vivax pLDH RDT (Bioline Malaria Ag Pf/Pf/Pv 05FK120; Standard Diagnostics, Gyeonhhi-do, South Korea) according to the manufacturer’s instructions.
DNA Extraction and Plasmodium Speciation
Each dried blood spot was processed into 3 discs from which genomic DNA was extracted by using QIAamp DNA Mini Kits and a QIAcube Robot (QIAGEN, Crawley, UK) according to the manufacturer’s instructions. DNA was eluted into a volume of 100 µL, and 10 µL was used in each PCR. An 18S rRNA gene-based multiplex PCR (20) was used to determine whether 4 human Plasmodium spp. were present in each sample.
Characterization of pfhrp2 and pfhrp3 Gene Deletions and Flanking Genes
Presence or absence of pfhrp2 (Pf3D7_0831800) and pfhrp3 (Pf3D7_1372200) genes was characterized by amplifying across exon1–exon2 and exon2 (7). Outcomes were classified as pfhrp2-positive or pfhrp3-positive (PCR result positive for exon1 and exon2 of the pfhrp2 or pfhrp3 gene) or pfhrp2-negative or pfhrp3-negative (PCR result negative for exon1 and exon2 of the pfhrp2 or pfhrp3 gene, but PCR positive result for 3 single-copy P. falciparum–specific genes [P. falciparum merozoite surface protein 1, P. falciparum merozoite surface protein 2, and P. falciparum glutmate-rich protein]) (21). Presence or absence of genes flanking pfhrp2 (Pf3D7_0831900/MAL7P1_230 and Pf3D7_0831700/MAL7P1_228) and pfhrp3 (Pf3D7_1372400/MAL12P1_485 and Pf3D7_1372100/MAL13P1_475) were characterized by PCR amplification of a fragment in each gene (7).
HRP2 Levels
HRP2 levels were measured for each sample by using Luminex mulitplex bead-based immunoassay described elsewhere (22). Dried blood spots were incubated overnight in elution buffer (0.05% phosphate-buffered saline, Tween 20, 0.05% NaN3) to give a 1:20 dilution of whole blood. A 50-µL elution was used in each assay. The threshold mean fluorescence intensity minus background (MFI – bg) signal that indicated true negativity for HRP2 was derived by testing 86 blood samples from a setting to which malaria was not endemic. The MFI – bg positivity cutoff value was the lognormal mean ± 3 SD for this malaria-negative population.
Microsatellite Analysis
We amplified 7 neutral microsatellite markers (TA1, PolyA, PfPK2, TA109, 2490, 313, and 383) from each sample (23) and assayed for size by using an ABI 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). We scored alleles manually by using Peak Scanner Software version 1.0 (Applied Biosystems), using a height of 300 relative fluorescence units as the minimal peak threshold.
Genetic Diversity and Population Genetic Analyses
We determined haplotypes for each parasite isolate from 7 microsatellite markers and used for genetic diversity and genetic relatedness analysis among parasites from Eritrea and between parasites from Eritrea and Peru (23) by using PHYLOViZ software (24). We calibrated sizes of 7 microsatellite markers against those of P. falciparum strain 3D7 before conducting genetic relatedness analysis, using a cutoff value of 2 (minimum differences for 2 loci) to compare parasites. We calculated expected heterozygosity (HE) values for each of the 7 microsatellite markers by using FSTAT software (https://www2.unil.ch/popgen/softwares/fstat.htm) and derived the mean HE for pfhrp2-positive and pfhrp2-negative parasite populations in Eritrea.
Statistical Analysis
We conducted graphics and statistical analysis by using GraphPad Prism 7.00 for Windows (GraphPad Software, La Jolla, CA, USA). Parasite densities were log-transformed for analysis and the geometric mean is reported. We used a Mann-Whitney test to compare log parasite densities between hospitals and between pfhrp2-positive and pfhrp2-negative parasites.
Ethics Considerations
The study was approved by the Eritrean MOH Research and Ethical Committees. The Northern Red Sea branch of the MOH and authorities of the 2 hospitals were informed, through an official letter, on the scope, coverage, and objectives of the study.
Patients were enrolled after providing consent following a detailed explanation about the investigation. Data from patients were recorded on structured forms. All patient specimens were given a unique identification number after collection, and only this number was used for data linkage.
Laboratory analyses of pfhrp2/pfhrp3 status of parasites and their genetic diversity and relatedness was conducted at the Australian Defence Force Malaria and Infectious Disease Institute. Analyses were approved by the Australian Defence Joint Health Command Low Risk Ethics Panel (LREP 15–004).
Patient Characteristics
A total of 51 patients recruited from the 2 hospitals participated in the study. We obtained characteristics for all of these patients (Table 1).
Microscopy and Plasmodium Speciation PCR
Microscopy at the NRL confirmed 50 infections only with P. falciparum and 1 infection only with P. vivax. These results were confirmed by PCR; no mixed infections or infections with other Plasmodium species were identified. The 1 infection with P. vivax was excluded from further analysis. The number of P. falciparum–infected patients was approximately equal between the hospitals, and patients from the 2 hospitals had comparable parasite densities (p = 0.47) (Table 1).
RDT
All 50 confirmed P. falciparum samples showed positive results for the pan-pLDH RDT, but only 19 showed positive results for the HRP2-based RDT. These samples showed an overall false-negative rate of 62.0%; a total of 21 (80.8%) of 26 at Ghindae Hospital and 10 (41.7%) of 24 at Massawa Hospital were false negative (Table 1).
Presence or Absence of pfhrp2- and pfhrp3-Negative Genes
At Ghindae Hospital, 80.8% (21/26) of samples contained pfhrp2-negative parasites and 92.3% (24/26) pfhrp3-negative parasites. At Massawa Hospital, 41.7% (10/24) of samples contained pfhrp2-negative parasites and 70.8% (17/24) pfhrp3-negative parasites (Table 2). When we combined sample data, 62.0% (31/50) of samples contained pfhrp2-negative parasites and 82.0% (41/50) pfhrp3-negative parasites. All pfhrp2-negative samples (31/50, 62.0%) were also pfhrp3 negative, but only 9 (18.0%) of 50 pfhrp3-negative samples were also pfhrp2-negative (Table 2; Figure 2).
Presence or Absence of Genes Flanking pfhrp2 and pfhrp3
All 31 pfhrp2-negative isolates had a deletion of the upstream gene but retained the downstream gene of pfhrp2. A total of 8 pfhrp2-positive samples had a deletion of the upstream gene. In contrast, of 41 pfhrp3-negative samples, 30 had deletions of both flanking genes, 2 had deletions of only the upstream gene, and 9 had deletions of only the downstream gene. We obtained different patterns of pfhrp2, pfhrp3, and their flanking gene status in samples collected from both hospitals (Table 2).
RDT Results and pfhrp2/pfhrp3
All 31 samples positive for P. falciparum by microscopy and PCR but negative by HRP2-based RDTs had deletions of pfhrp2 and pfhrp3. Conversely, all 19 pfhrp2-positive samples showed positive results by HRP2-based RDTs, including 12 pfhrp3-negative samples (Figure 2).
Parasite Density and pfhrp2/pfhrp3
We determined the relationship between parasite density and pfhrp2/pfhrp3 status (Figure 2). The geometric mean parasite density for the 5 pfhrp2-positive patients at Ghindae Hospital was 11,069 parasites/µL (95% CI 4,886–25,074 parasites/µL), which is comparable with that for the 21 pfhrp2-negative patients (6,111 parasites/µL, 95% CI 2,664–14,020 parasites/µL; p = 0.86). The geometric mean parasite density for the 14 pfhrp2-positive patients at Massawa Hospital was 2,999 parasites/µL (95% CI 1,113–8,080 parasites/µL), which was significantly lower than that for the 10 pfhrp2-negative patients (11,783 parasites/µL, 95% CI 4,695–29,568 parasites/µL; p = 0.021). When data for patients from the 2 hospitals were combined, geometric mean parasite densities in pfhrp2-positive and pfhrp2-negative patients were not significantly different (p = 0.085).
HRP2 Levels and pfhrp2/pfhrp3
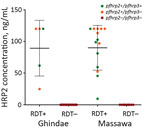
HRP2 levels, as indicated by mean MFI – bg values, were 18,885 (95% CI 16,061–21,710) for 19 pfhrp2-positive samples and 15.2 (95% CI 9.9–20.4) for 31 pfhrp2-negative samples. We found no significant difference between hospitals (p>0.05). When MFI – bg values were converted to HRP2 concentrations, pfhrp2-negative samples had an undetectable amount of HRP2, and pfhrp2-positive samples had an HRP level >9.5 ng/mL (Figure 3).
Genetic Diversity of Parasites
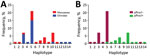
We determined 14 unique haplotypes for 50 samples on the basis of 7-loci microsatellite genotyping (Figure 4, panel A). All samples contained only 1 dominant haplotype. Nine haplotypes were detected for 19 pfhrp2-positive samples, and the maximum number of isolates sharing 1 haplotype (H10) was 7. Six haplotypes were detected for 31 pfhrp2-negative samples. Different haplotypes were observed for pfhrp2-negative and pfhrp2-positive populations except for haplotype H5, which was present in 20 pfhrp2-negative/pfhrp3-negative isolates and 1 pfhrp2-positive/pfhrp3-negative isolate (Figure 4, panel A). The 3 dominant haplotypes (H3, H5, and H10) were detected at both hospitals (Figure 4, panel B).
HE values were 0.44 for pfhrp2-positive parasite populations and 0.11 for pfhrp2-negative parasite populations. These values indicated an overall lower level of genetic diversity in the pfhrp2-negative parasite population. Three of the 7 markers in pfhrp2-negative parasites had HE values of 0, indicating no diversity, but only 1 marker in pfhrp2-positive parasites had an HE value of 0.
Genetic Relatedness of Parasites
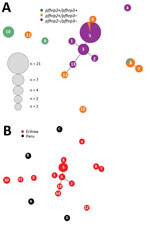
Of 9 haplotypes for pfhrp2-positive parasites from Eritrea, 4 (H9, H10, H11, and H12) were scattered and 2 (H7 and H8) were genetically closely related (Figure 5, panel A). In contrast, for pfhrp2-negative parasites, 5 of the 6 haplotypes (H1, H2, H3, H5, and H13) were genetically closely related and formed a cluster. Also linked to this cluster were haplotypes H6 and H14, which were pfhrp2 positive/pfhrp3 negative. We found 1 unrelated pfhrp2-negative haplotype (H4) outside the major cluster, which indicated different genetic lineages (Figure 5, panel A). There were 2 clusters for pfhrp3-negative parasites.
The 14 haplotypes observed in Eritrea were unrelated to any of the 5 haplotypes observed in Peru. This finding suggested distant genetic lineages between isolates from these 2 countries (Figure 5, panel B).
Since malaria RDTs became available in the 1990s, growth in the number of tests, manufacturers, and volumes sold has been exponential (25,26). In parallel, the proportion of fever case-patients having access to diagnostic services before treatment has also expanded, particularly in Africa, largely attributed to the implementation of HRP2-based malaria RDTs (3). To continue reducing malaria transmission, use of RDTs must be expanded further, and tests must remain affordable, accurate, and user friendly.
Many factors can affect the accuracy of RDTs, and each factor should be investigated as a potential cause of false-negative results (27). In this instance, the Eritrea MOH had already investigated reports of false-negative results for RDTs and ruled out several possible causes (6). In our report, molecular and protein characterizations of prospectively collected specimens provided unambiguous evidence that confirmed that incidences of false-negative results in Eritrea were caused by a high prevalence of parasites having deletions of the pfhrp2 and pfhrp3 genes.
Our data showed that, in March 2016, a total of 80.8% (21/26) patients at Ghindae Hospital and 41.7% (10/24) at Massawa Hospital were infected with pfhrp2-negative parasites. All of these patients had undetectable levels of HRP2 in blood samples, and all showed negative results by HRP2-based RDTs and positive results by pan-pLDH–based RDTs. All samples were confirmed by microscopy and PCR as having only P. falciparum infections. Conversely, the remaining patient samples were pfhrp2 positive, and all showed positive results for HRP2-based and pan-pLDH–based RDTs, and had HRP2 concentrations >9.5 ng/mL. These data confirmed that false-negative RDT results were caused by parasites lacking pfhrp2.
Although pfhrp2-negative parasites have been increasingly reported in several countries in South America, Asia, and Africa, they were mostly sporadic and showed low prevalences, except in the Amazon region of South America. Our study showed a high prevalence of parasites lacking pfhrp2 that caused false-negative RDT results outside South America and a 100% correlation between parasites lacking the pfhrp2 gene, the HRP2 antigen, and false-negative RDT results. These pfhrp2 gene deletions were detected after investigations triggered by field reports of false-negative RDT results for symptomatic patients and categorically confirmed the role of this issue in malaria case management.
We showed correlations between patient demographics, parasite densities, and parasite pfhrp2 gene status at the individual level. Our data showed that P. falciparum infections occurred across a range of parasite densities and age groups. All but 2 patients were symptomatic over the 2 weeks before testing, regardless of being infected with pfhrp2-positive or pfhrp2-negative parasites; the remaining 2 patients were infected with pfhrp2-negative parasites. Geometric mean parasite densities for pfhrp2-negative patients were comparable to those for pfhrp2-positive patients in this sample set. Further studies with larger sample sizes are required to confirm this finding.
Another useful finding of our study was the high prevalence of pfhrp3 gene deletions in samples from Eritrea, which resulted in a high prevalence of parasites with concomitant pfhrp2 and pfhrp3 gene deletions. Every sample that was pfhrp2-negative was also pfhrp3-negative, which showed that 2 genes on different chromosomes were deleted in parallel. It appears that the dual-deleted parasites (pfhrp2 negative/pfhrp3-negative) form clusters, and pfhrp2-positive/pfhrp3-negative parasites are scattered outside these clusters, suggesting that these parasites might have evolved from different genetic backgrounds. Although pfhrp3 status did not affect results of RDTs used in this study, this status could affect results of other RDTs because pfhrp3 shares sequence similarity with pfhrp2, and cross-reactivity between HRP2 and HRP3 has been reported for some RDTs (28). It is clear that regardless of the brand of HRP2-based RDTs used, parasites with dual pfhrp2 and pfhrp3 deletions will cause failure of RDTs because there is no possibility for cross-reactivity between HRP3 and HRP2 to occur.
A major issue is whether pfhrp2-negative parasites emerged locally and what is the main driving force behind their prevalence. In Peru, pfhrp2-negative parasites were detected in 4 of 5 major parasite populations during the late 1990s, and their prevalence increased in the absence of HRP2-based RDT pressure because microscopy was the primary diagnostic test used in that country (23). Spread of these parasites in Peru might have been driven by an undefined biological advantage associated with deletion of the pfhrp2 gene.
In Eritrea, low genetic diversity of pfhrp2-negative parasites compared with that of pfhrp2-positive parasites and presence of a closely related cluster consisting of all but 1 pfhrp2-negative parasite suggest that clonal expansion of pfhrp2-negative parasites was probably caused by selection by use of HRP2-based RDTs. Because HRP2-based RDTs have been widely used in Eritrea since 2006, compliance with the recommended test before treatment is universally applied, and treatment adherence is high, conditions are ideal for selection of pfhrp2-negative parasites. This selection is predicted by recent mathematical modeling, which showed that exclusive use of HRP2-based RDTs exerts strong selection pressure for pfhrp2-negative parasites in communities and that HRP2/P. falciparum pan-LDH combination RDTs do not provide selection pressure for pfhrp2-negative parasites (18). Furthermore, low malaria prevalence in Eritrea might have also contributed to selection of pfhrp2-negative parasites once they emerge. However, historical samples were not available for a retrospective analysis of the dynamics and evolution of the pfhrp2-negative parasites in Eritrea.
The presence of a unique pfhrp2-negative parasite outside the main cluster suggests that the pfhrp2 deletion has occurred at least twice in parasites in Eritrea. The pfhrp2-negative parasites in Eritrea and Peru showed distinct haplotypes, strongly suggesting de novo development of these parasites in both locations. This suggestion would imply that all malaria-endemic areas are at risk and that there is an urgent need to map the prevalence of pfhrp2-negative parasites to inform case management policy. The immediate mapping priority should be for countries/areas near Eritrea. It is also critical for research and development and improvements in alternative biomarkers of P. falciparum. Currently, there is only 1 P. falciparum and 1 P. falciparum/P. vivax–detecting RDT specific for pLDH that meet WHO procurement criteria. Most manufacturers have difficulties in producing RDTs that can consistently detect P. falciparum at a concentration of 200 parasites/µL by using antibodies against P. falciparum–pLDH (29).
Emergence of pfhrp2-negative parasites poses a major threat to malaria control programs because patients infected with these parasites are not given a correct diagnosis and treatment. Because of broad use of HRP2-based RDTs, particularly in Africa, the magnitude of selection pressure and potential scope of the problem are large. Confirming pfhrp2/3 deletions as the cause of false-negative RDT results requires multiple PCRs performed by experienced technicians. Guidance on investigating pfhrp2 deletions and accurate reporting is available (21). In addition, WHO has recently released a standard survey tool to determine whether pfhrp2/3 gene deletions in P. falciparum isolated from patients with confirmed symptomatic malaria have reached a threshold triggering a change in diagnostic strategy (30).
In conclusion, we confirmed that frequently reported false-negative results for P. falciparum in specimens tested by using HRP2-based RDTs in Eritrea were caused by high proportions of parasites with deletions of pfhrp2/pfhrp3 genes. It is clear that HRP2-based RDTs are no longer appropriate diagnostic tools for malaria case management in this country. RDTs detecting other parasite antigens, such as P. falciparum–pLDH, have been implemented. Our results show the need to seriously investigate reports of false-negative RDT results and conduct surveys to determine the prevalence of pfhrp2/3 deletions in neighboring regions. International technical guidance is also needed to assist countries in planning pfhrp2/3 surveillance activities and laboratory investigations and adapting procurement and case management strategies when pfhrp2/3 gene deletions are confirmed.
Dr. Berhane is director of the Communicable Diseases Control Division, Ministry of Health, Asmara, Eritrea. His primary research interest is control of communicable diseases, including malaria.
Acknowledgments
We thank all study subjects for participating in the study.
Parasite genetic analysis was supported by the US Department of Defense Global Emerging Infections Surveillance N.C.W., S.D., Q.C., K.A., and K.G. were supported by a grant from the US Department of Defense Global Emerging Infections Surveillance J.C. was supported by a grant from UNITAID.
A.B., S. Mihreteab, S. Mohamed, F.H., G.E., A.C., and A.Z. conducted hospital surveys and performed training and sample collection; K.A. performed molecular characterization of pfhrp2/pfhrp3 and flanking genes; K.G. and S.D. performed microsatellite typing and genetic analyses; N.C.W. and Q.C. performed molecular supervision; E.R. and V.U. performed antigen detection; K.G., Q.C., and M.L.G. analyzed data; A.B., S. Mihreteab, E.R., Q.C., and J.C. wrote the manuscript; and J.C. coordinated the study.
References
- Nyarango PM, Gebremeskel T, Mebrahtu G, Mufunda J, Abdulmumini U, Ogbamariam A, et al. A steep decline of malaria morbidity and mortality trends in Eritrea between 2000 and 2004: the effect of combination of control methods. Malar J. 2006;5:33. DOIPubMedGoogle Scholar
- Barat LM. Four malaria success stories: how malaria burden was successfully reduced in Brazil, Eritrea, India, and Vietnam. Am J Trop Med Hyg. 2006;74:12–6.PubMedGoogle Scholar
- World Health Organization. World malaria report 2016 [cited 2017 Nov 29]. http://www.who.int/malaria/publications/world-malaria-report-2016/report/en/
- Berhane A. Changing pettern of P. vivax malaria in Eritrea: implication for the national malaria control. Journal of the Eritrean Medical Association. 2011;1:17–20.
- World Health Organization. WHO Global Malaria Programme Information note on recommended selection criteria for procurement of malaria rapid diagnostic tests (RDTs), 2012 [cited 2017 Nov 29]. http://www.who.int/malaria/publications/atoz/rdt-selection-criteria.pdf
- Berhane A, Russom M, Bahta I, Hagos F, Ghirmai M, Uqubay S. Rapid diagnostic tests failing to detect Plasmodium falciparum infections in Eritrea: an investigation of reported false negative RDT results. Malar J. 2017;16:105. DOIPubMedGoogle Scholar
- Gamboa D, Ho MF, Bendezu J, Torres K, Chiodini PL, Barnwell JW, et al. A large proportion of P. falciparum isolates in the Amazon region of Peru lack pfhrp2 and pfhrp3: implications for malaria rapid diagnostic tests. PLoS One. 2010;5:e8091. DOIPubMedGoogle Scholar
- Murillo Solano C, Akinyi Okoth S, Abdallah JF, Pava Z, Dorado E, Incardona S, et al. Deletion of Plasmodium falciparum histidine-rich protein 2 (pfhrp2) and histidine-rich protein 3 (pfhrp3) genes in Colombian parasites. PLoS One. 2015;10:e0131576. DOIPubMedGoogle Scholar
- Akinyi Okoth S, Abdallah JF, Ceron N, Adhin MR, Chandrabose J, Krishnalall K, et al. Variation in Plasmodium falciparum histidine-rich protein 2 (pfhrp2) and Plasmodium falciparum histidine-rich protein 3 (pfhrp3) gene deletions in Guyana and Suriname. PLoS One. 2015;10:e0126805. DOIPubMedGoogle Scholar
- Rachid Viana GM, Akinyi Okoth S, Silva-Flannery L, Lima Barbosa DR, Macedo de Oliveira A, Goldman IF, et al. Histidine-rich protein 2 (pfhrp2) and pfhrp3 gene deletions in Plasmodium falciparum isolates from select sites in Brazil and Bolivia. PLoS One. 2017;12:e0171150. DOIPubMedGoogle Scholar
- Maltha J, Gamboa D, Bendezu J, Sanchez L, Cnops L, Gillet P, et al. Rapid diagnostic tests for malaria diagnosis in the Peruvian Amazon: impact of pfhrp2 gene deletions and cross-reactions. PLoS One. 2012;7:e43094. DOIPubMedGoogle Scholar
- Bharti PK, Chandel HS, Ahmad A, Krishna S, Udhayakumar V, Singh N. Prevalence of pfhrp2 and/or pfhrp3 gene deletion in Plasmodium falciparum population in eight highly endemic states in India. PLoS One. 2016;11:e0157949. DOIPubMedGoogle Scholar
- Li P, Xing H, Zhao Z, Yang Z, Cao Y, Li W, et al. Genetic diversity of Plasmodium falciparum histidine-rich protein 2 in the China-Myanmar border area. Acta Trop. 2015;152:26–31. DOIPubMedGoogle Scholar
- Koita OA, Doumbo OK, Ouattara A, Tall LK, Konaré A, Diakité M, et al. False-negative rapid diagnostic tests for malaria and deletion of the histidine-rich repeat region of the hrp2 gene. Am J Trop Med Hyg. 2012;86:194–8. DOIPubMedGoogle Scholar
- Wurtz N, Fall B, Bui K, Pascual A, Fall M, Camara C, et al. Pfhrp2 and pfhrp3 polymorphisms in Plasmodium falciparum isolates from Dakar, Senegal: impact on rapid malaria diagnostic tests. Malar J. 2013;12:34. DOIPubMedGoogle Scholar
- Parr JB, Verity R, Doctor SM, Janko M, Carey-Ewend K, Turman BJ, et al. Pfhrp2-deleted Plasmodium falciparum parasites in the Democratic Republic of Congo: a national cross-sectional survey. J Infect Dis. 2017;216:36–44.PubMedGoogle Scholar
- Amoah LE, Abankwa J, Oppong A. Plasmodium falciparum histidine rich protein-2 diversity and the implications for PfHRP 2: based malaria rapid diagnostic tests in Ghana. Malar J. 2016;15:101. DOIPubMedGoogle Scholar
- Gatton ML, Dunn J, Chaudhry A, Ciketic S, Cunningham J, Cheng Q. Implications of parasites lacking Plasmodium falciparum histidine-rich protein 2 on malaria morbidity and control when rapid diagnostic tests are used for diagnosis. J Infect Dis. 2017;215:1156–66. DOIPubMedGoogle Scholar
- World Health Organization Western Pacific Region. Malaria microscopy standard operating procedures, 2016 [cited 2017 Nov 29]. http://www.wpro.who.int/mvp/lab_quality/mm_sop/en/
- Padley D, Moody AH, Chiodini PL, Saldanha J. Use of a rapid, single-round, multiplex PCR to detect malarial parasites and identify the species present. Ann Trop Med Parasitol. 2003;97:131–7. DOIPubMedGoogle Scholar
- Cheng Q, Gatton ML, Barnwell J, Chiodini P, McCarthy J, Bell D, et al. Plasmodium falciparum parasites lacking histidine-rich protein 2 and 3: a review and recommendations for accurate reporting. Malar J. 2014;13:283. DOIPubMedGoogle Scholar
- Rogier E, Plucinski M, Lucchi N, Mace K, Chang M, Lemoine JF, et al. Bead-based immunoassay allows sub-picogram detection of histidine-rich protein 2 from Plasmodium falciparum and estimates reliability of malaria rapid diagnostic tests. PLoS One. 2017;12:e0172139. DOIPubMedGoogle Scholar
- Akinyi S, Hayden T, Gamboa D, Torres K, Bendezu J, Abdallah JF, et al. Multiple genetic origins of histidine-rich protein 2 gene deletion in Plasmodium falciparum parasites from Peru. Sci Rep. 2013;3:2797. DOIPubMedGoogle Scholar
- Ribeiro-Gonçalves B, Francisco AP, Vaz C, Ramirez M, Carriço JA. PHYLOViZ Online: web-based tool for visualization, phylogenetic inference, analysis and sharing of minimum spanning trees. Nucleic Acids Res. 2016;44(W1):W246-51. DOIPubMedGoogle Scholar
- Incardona S, Serra-Casas E, Champouillon N, Nsanzabana C, Cunningham J, González IJ. Global survey of malaria rapid diagnostic test (RDT) sales, procurement and lot verification practices: assessing the use of the WHO-FIND Malaria RDT Evaluation Programme (2011-2014). Malar J. 2017;16:196. DOIPubMedGoogle Scholar
- World Health Organization. Malaria rapid diagnostic test performance: summary results of WHO product testing of malaria RDTs: round 1–7 (2008–2016), 2017 [cited 2017 Nov 29] http://www.who.int/malaria/publications/atoz/978924151268/en/
- World Health Organization. False-negative RDT results and implications of new reports of P. falciparum histidine-rich protein 2/3 gene deletions, 2016 [cited 2017 Nov 29] http://www.who.int/malaria/publications/atoz/information-note-hrp2-based-rdt/en/
- Baker J, McCarthy J, Gatton M, Kyle DE, Belizario V, Luchavez J, et al. Genetic diversity of Plasmodium falciparum histidine-rich protein 2 (PfHRP2) and its effect on the performance of PfHRP2-based rapid diagnostic tests. J Infect Dis. 2005;192:870–7. DOIPubMedGoogle Scholar
- World Health Organization. Malaria rapid diagnostic test performance: results of WHO product testing of malaria RDTs: round 7 (2015–2016), 2017 [cited 2017 Nov 29]. http://www.who.int/malaria/publications/atoz/978924151268
- World Health Organization. Determining whether histidine rich protein gene deletions causing negative HRP2 RDT results among symptomatic falciparum patients have reached a threshold for switching malaria RDTs, 2017 [cited 2017 Nov 29]. http://www.who.int/malaria/areas/diagnosis/en/
Figures
Tables
Cite This ArticleTable of Contents – Volume 24, Number 3—March 2018
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Qin Cheng, Drug Resistance and Diagnostics, Australian Defence Force Malaria and Infectious Disease Institute, Weary Dunlop Dr, Gallipoli Barracks, Enoggera, Brisbane, Queensland 4051, Australia:
Top