Volume 24, Number 6—June 2018
Research
Genomic Epidemiology of Global Carbapenemase-Producing Enterobacter spp., 2008–2014
Figure 4
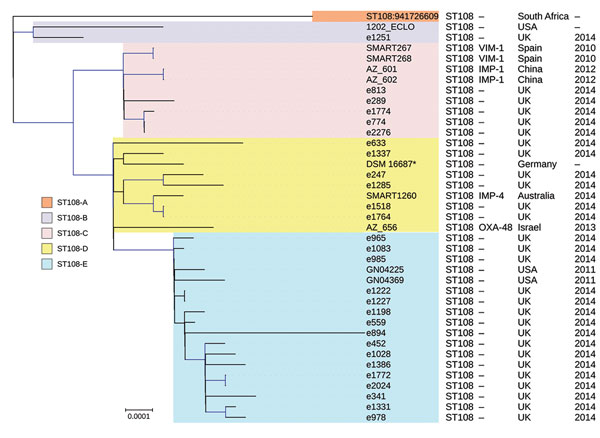
Figure 4. Phylogenetic tree of the different clades among 39 Enterobacter hormaechei subsp. oharae ST108 isolates identified from Enterobacter spp. isolates collected in the Merck Study for Monitoring Antimicrobial Resistance Trends, 2008–2014, and the AstraZeneca global surveillance program, 2012–2014. The tree was rooted with E. hormaechei subsp. hormaechei isolate ATCC49162. A total of 317,867 core single-nucleotide polymorphisms were found; 27,705 were used to draw the tree (after phages and recombination sites were excluded). The isolates from other studies were negative for carbapenemases. Clades are grouped by color. KPC, Klebsiella pneumoniae carbapenemase; NDM, New Delhi metallo-β-lactamase; OXA, oxacillin; ST, sequence type; –, information missing. Scale bar indicates nucleotide substitutions per site.
1These co-authors contributed equally to this article.
2Current affiliation: Jackson Laboratory for Genomic Medicine, Farmington, Connecticut, USA.