Volume 25, Number 11—November 2019
Dispatch
Isolation of Legionella pneumophila by Co-culture with Local Ameba, Canada
Figure 2
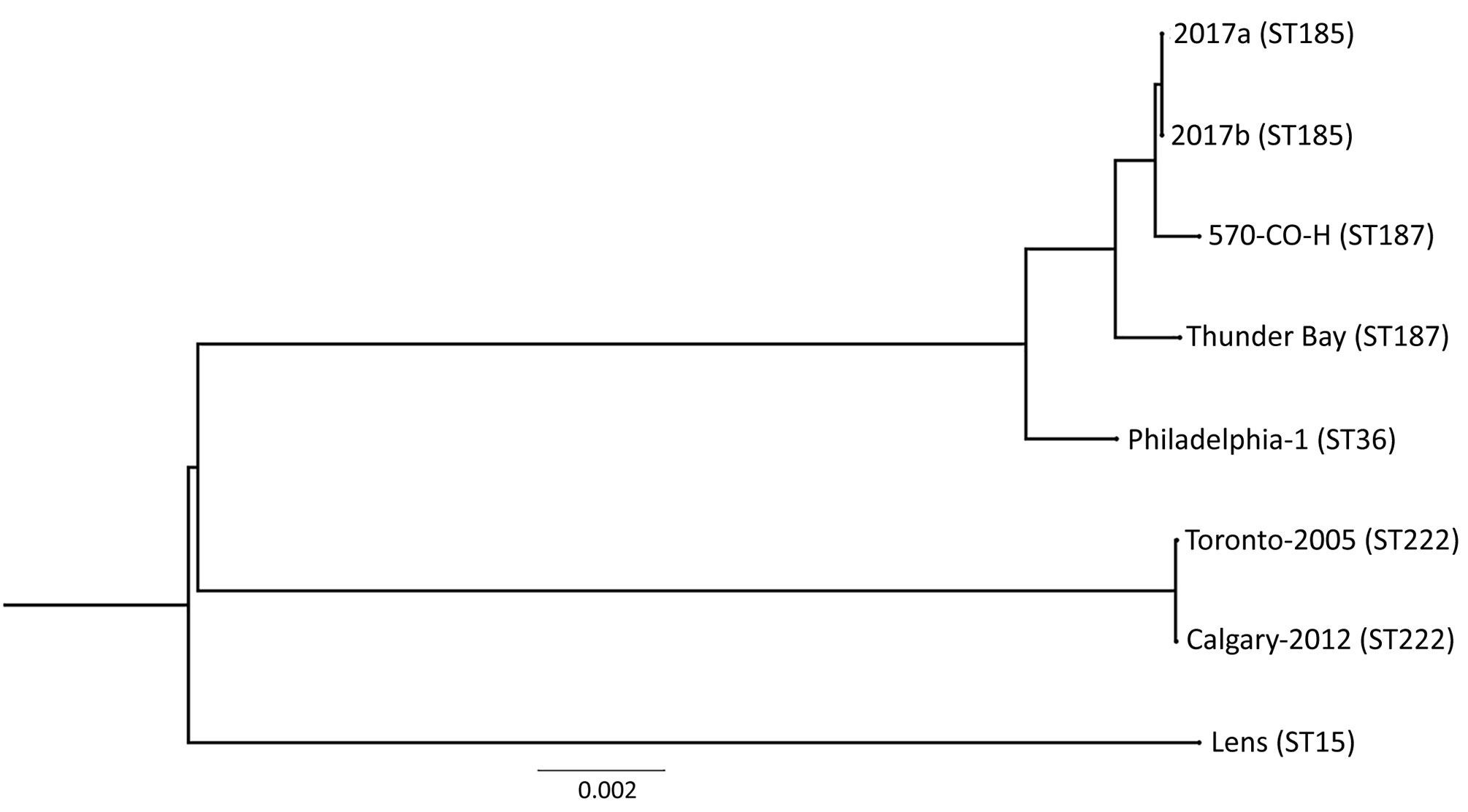
Figure 2. Phylogenetic tree depicting the relationship between Legionella pneumophila isolates identified during investigation of legionellosis in an immunocompromised 3-year-old girl, Calgary, Alberta, Canada, and reference sequences. L. pneumophila core ortholog-based maximum-likelihood phylogenetic tree shows 8 previously published genomes and sequences of the 2 isolates from this study (2017a, clinical isolate from patient; 2017b, environmental isolate from hot tub in patient’s home). Tree construction was performed by using 2,403 orthologous sequences (2,471,034 nt). Each ortholog sequence was independently aligned with the MUSCLE algorithm (https://www.ebi.ac.uk/Tools/msa/muscle) and concatenated into a single superalignment, which then was subjected to 1,000 bootstrap iterations to find the maximum-likelihood phylogeny. Scale bar indicates the number of nucleotide substitutions per site. ST, sequence type.