Sand Fly–Associated Phlebovirus with Evidence of Neutralizing Antibodies in Humans, Kenya
David P. Tchouassi

, Marco Marklewitz, Edith Chepkorir, Florian Zirkel
1, Sheila B. Agha, Caroline C. Tigoi, Edith Koskei, Christian Drosten, Christian Borgemeister, Baldwyn Torto, Sandra Junglen
2
, and Rosemary Sang
2
Author affiliations: International Centre of Insect Physiology and Ecology, Nairobi, Kenya (D.P. Tchouassi, E. Chepkorir, S.B. Agha, C.C. Tigoi, B. Torto, R. Sang); Charité-Universitätsmedizin Berlin, Berlin, Germany (M. Marklewitz, F. Zirkel, C. Drosten, S. Junglen); German Center for Infection Research, Berlin (M. Marklewitz, F. Zirkel, C. Drosten, S. Junglen); Center for Virus Research, Kenya Medical Research Institute, Nairobi (E. Koskei, R. Sang); University of Bonn, Bonn, Germany (C. Borgemeister)
Main Article
Figure 2
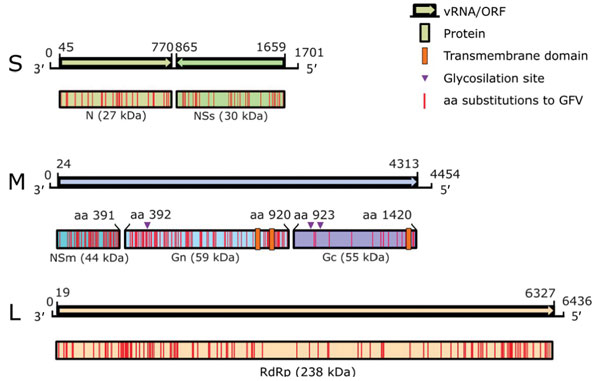
Figure 2. Genome organization of novel sand fly–associated phlebovirus Ntepes virus identified in Kenya. Sequence length of the L, M, and S segments (in bp) and encoded predicted proteins RdRp, Gn, Gc, N, and nonstructural proteins NSm and NSs (in kDa) are indicated; ORF positions (length in bp) are also indicated. GFV, Gabek Forest virus; L, large segment (encoding the RdRp protein); M, medium segment (encoding the nonstructural protein NSm and the 2 glycoproteins Gn and Gc); N, nucleocapsid protein; ORF, open reading frame; RdRp, RNA-dependent RNA polymerase; S, small segment (encoding the N protein and nonstructural protein NSs in an ambisense manner); vRNA, virus RNA.
Main Article
Page created: March 17, 2019
Page updated: March 17, 2019
Page reviewed: March 17, 2019
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
