Volume 25, Number 4—April 2019
Research
Human-Origin Influenza A(H3N2) Reassortant Viruses in Swine, Southeast Mexico
Figure 4
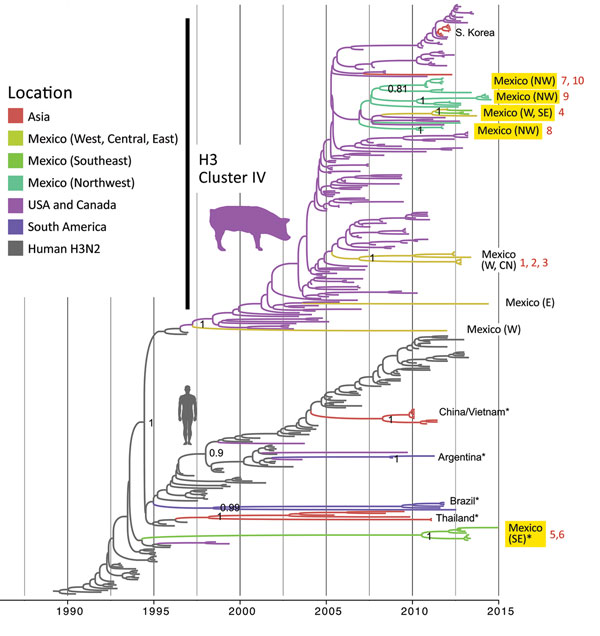
Figure 4. Evolutionary relationships between H3 segments of avian influenza viruses collected in humans and swine globally. Time-scaled Bayesian maximum clade credibility tree is inferred for the H3 segment. The tree includes newly generated sequences from northwest and southeast Mexico, along with background sequences from swine in Mexico, Asia, the United States, and Canada, as well as seasonal H3N2 viruses from humans. The color of each branch indicates the most probable location state. Posterior probabilities for key nodes are provided. Clades with viruses from swine in Mexico obtained for this study are highlighted in yellow. *Major virus introductions into a location, indicating direct introductions from humans. CN, central; NW, northwest; S, south; SE, southeast; W, west. The 10 H3N2 viruses selected for antigenic characterization (Figure 6) are indicated by numbers 1–10 in red. A more detailed phylogeny, including tip labels and all posterior probabilities, is provided in the Appendix Figure.
1Current affiliation: Kansas State University, Manhattan, Kansas, USA.