Volume 25, Number 4—April 2019
Research
Human-Origin Influenza A(H3N2) Reassortant Viruses in Swine, Southeast Mexico
Figure 8
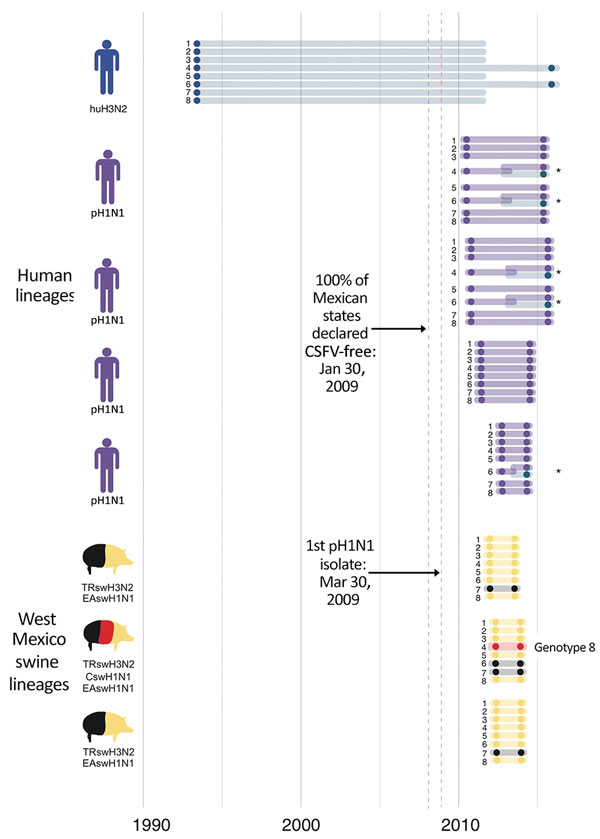
Figure 8. Introductions of influenza A viruses into swine in southeast Mexico. Introductions of these viruses from humans and swine into southeast Mexico are indicated as groups of 8 horizontal lines, and each line represents 1 of 8 segments of the virus genome. Segments are numbered 1–8, longest to shortest, according to convention: 1, polymerase basic 2; 2, polymerase basic 1; 3, polymerase acidic; 4, hemagglutinin; 5, nucleoprotein; 6, neuraminidase; 7, matrix protein; 8, nonstructural. The position of each shaded circle along the x-axis represents the estimated timing of virus introduction, estimated directly from the maximum clade credibility trees. Lines and circles are shaded according to the virus lineage, similar to that in Figure 2. *Reassortment events. Introduction of genotype 8 viruses is indicated. The blue dotted vertical line represents the date of the declaration of Secretariat of Agriculture, Livestock, Rural Development, Fisheries and Food in Mexico that classical swine fever virus had been eradicated from all states in Mexico during January 2009. The red dotted vertical line represents the date of first isolation of pH1N1 virus in humans. CSFV, classical swine fever virus; CswH1, classical swine H1N1; EAswH1, avian-like Eurasian swine H1N1; huH3N2, human H3N2; pH1N1, pandemic H1N1 clade; TRswH3N2, triple reassortant swine H3N2.
1Current affiliation: Kansas State University, Manhattan, Kansas, USA.