Volume 25, Number 4—April 2019
Research
Human-Origin Influenza A(H3N2) Reassortant Viruses in Swine, Southeast Mexico
Figure 6
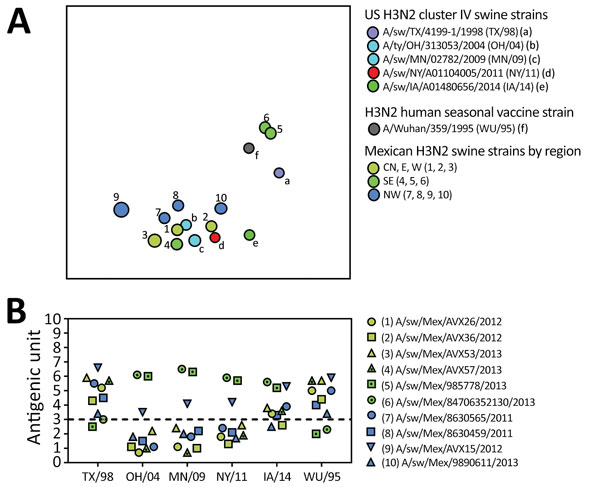
Figure 6. Antigenic relationships between contemporary influenza A (H3N2) viruses from Mexico and representative strains from the United States. A) Antigenic map of contemporary swine H3N2 viruses from Mexico and the United States. Antigenic clusters are indicated by color as in Figures 4 and 5. CN, central; E, east; NW, northwest; SE, southeast; W, west. B) Antigenic distance between H3N2 virus from Mexico and representative swine strains from the United States and a putative ancestral human seasonal vaccine virus strain (WU/95). The US swine strains represent antigenic clusters previously defined as TX/98 (cornflower), OH/04 and MN/09 (cyan), NY/11 (red), and IA/14 (green) (27,31).
References
- Mena I, Nelson MI, Quezada-Monroy F, Dutta J, Cortes-Fernández R, Lara-Puente JH, et al. Origins of the 2009 H1N1 influenza pandemic in swine in Mexico. eLife. 2016;5:e16777. DOIPubMedGoogle Scholar
- Koen J. A practical method for field diagnosis of swine diseases. Am J Vet Med. 1919;14:468–70.
- Anderson TK, Macken CA, Lewis NS, Scheuermann RH, Van Reeth K, Brown IH, et al. A phylogeny-based global nomenclature system and automated annotation tool for H1 hemagglutinin genes from swine influenza viruses. mSphere. 2016;1:pii: e00275–16.
- Vijaykrishna D, Smith GJD, Pybus OG, Zhu H, Bhatt S, Poon LLM, et al. Long-term evolution and transmission dynamics of swine influenza A virus. Nature. 2011;473:519–22. DOIPubMedGoogle Scholar
- Watson SJ, Langat P, Reid SM, Lam TT, Cotten M, Kelly M, et al.; ESNIP3 Consortium. ESNIP3 Consortium. Molecular epidemiology and evolution of influenza viruses circulating within European swine between 2009 and 2013. J Virol. 2015;89:9920–31. DOIPubMedGoogle Scholar
- Lam TT, Zhu H, Wang J, Smith DK, Holmes EC, Webster RG, et al. Reassortment events among swine influenza A viruses in China: implications for the origin of the 2009 influenza pandemic. J Virol. 2011;85:10279–85. DOIPubMedGoogle Scholar
- Nelson MI, Viboud C, Vincent AL, Culhane MR, Detmer SE, Wentworth DE, et al. Global migration of influenza A viruses in swine. Nat Commun. 2015;6:6696. DOIPubMedGoogle Scholar
- Takemae N, Parchariyanon S, Damrongwatanapokin S, Uchida Y, Ruttanapumma R, Watanabe C, et al. Genetic diversity of swine influenza viruses isolated from pigs during 2000 to 2005 in Thailand. Influenza Other Respi Viruses. 2008;2:181–9. DOIPubMedGoogle Scholar
- Nelson MI, Lemey P, Tan Y, Vincent A, Lam TT, Detmer S, et al. Spatial dynamics of human-origin H1 influenza A virus in North American swine. PLoS Pathog. 2011;7:e1002077. DOIPubMedGoogle Scholar
- Vargas Terán M, Calcagno Ferrat N, Lubroth J. Situation of classical swine fever and the epidemiologic and ecologic aspects affecting its distribution in the American continent. Ann N Y Acad Sci. 2004;1026:54–64. DOIPubMedGoogle Scholar
- Slomka MJ, Densham ALE, Coward VJ, Essen S, Brookes SM, Irvine RM, et al. Real time reverse transcription (RRT)-polymerase chain reaction (PCR) methods for detection of pandemic (H1N1) 2009 influenza virus and European swine influenza A virus infections in pigs. Influenza Other Respi Viruses. 2010;4:277–93. DOIPubMedGoogle Scholar
- Zhang J, Gauger PC. Isolation of swine influenza virus in cell cultures and embryonated chicken eggs. Methods Mol Biol. 2014;1161:265–76. DOIPubMedGoogle Scholar
- Zhang J, Harmon KM. RNA extraction from swine samples and detection of influenza A virus in swine by real-time RT-PCR. Methods Mol Biol. 2014;1161:277–93. DOIPubMedGoogle Scholar
- Nelson M, Culhane MR, Rovira A, Torremorell M, Guerrero P, Norambuena J. Novel human-like influenza A viruses circulate in swine in Mexico and Chile. PLoS Curr. 2015;7:7.PubMedGoogle Scholar
- Hoffmann E, Stech J, Guan Y, Webster RG, Perez DR. Universal primer set for the full-length amplification of all influenza A viruses. Arch Virol. 2001;146:2275–89. DOIPubMedGoogle Scholar
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. DOIPubMedGoogle Scholar
- Bao Y, Bolotov P, Dernovoy D, Kiryutin B, Zaslavsky L, Tatusova T, et al. The influenza virus resource at the National Center for Biotechnology Information. J Virol. 2008;82:596–601. DOIPubMedGoogle Scholar
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–90. DOIPubMedGoogle Scholar
- Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29:1969–73. DOIPubMedGoogle Scholar
- Suchard MA, Rambaut A. Many-core algorithms for statistical phylogenetics. Bioinformatics. 2009;25:1370–6. DOIPubMedGoogle Scholar
- Minin VN, Suchard MA. Counting labeled transitions in continuous-time Markov models of evolution. J Math Biol. 2008;56:391–412. DOIPubMedGoogle Scholar
- Lemey P, Rambaut A, Bedford T, Faria N, Bielejec F, Baele G, et al. Unifying viral genetics and human transportation data to predict the global transmission dynamics of human influenza H3N2. PLoS Pathog. 2014;10:e1003932. DOIPubMedGoogle Scholar
- R Core Team. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2012 [cited 2018 Dec 19]. http://www.r-project.org/.
- Lewis NS, Anderson TK, Kitikoon P, Skepner E, Burke DF, Vincent AL. Substitutions near the hemagglutinin receptor-binding site determine the antigenic evolution of influenza A H3N2 viruses in U.S. swine. J Virol. 2014;88:4752–63. DOIPubMedGoogle Scholar
- Kitikoon P, Gauger PC, Vincent AL. Hemagglutinin inhibition assay with swine sera. Methods Mol Biol. 2014;1161:295–301. DOIPubMedGoogle Scholar
- Nelson MI, Wentworth DE, Culhane MR, Vincent AL, Viboud C, LaPointe MP, et al. Introductions and evolution of human-origin seasonal influenza a viruses in multinational swine populations. J Virol. 2014;88:10110–9. DOIPubMedGoogle Scholar
- Abente EJ, Santos J, Lewis NS, Gauger PC, Stratton J, Skepner E, et al. The molecular determinants of antibody recognition and antigenic drift in the H3 hemagglutinin of swine influenza A virus. J Virol. 2016;90:8266–80. DOIPubMedGoogle Scholar
- Beaudoin A, Johnson S, Davies P, Bender J, Gramer M. Characterization of influenza a outbreaks in Minnesota swine herds and measures taken to reduce the risk of zoonotic transmission. Zoonoses Public Health. 2012;59:96–106. DOIPubMedGoogle Scholar
- Sandbulte MR, Spickler AR, Zaabel PK, Roth JA. Optimal use of vaccines for control of influenza A virus in swine. Vaccines (Basel). 2015;3:22–73. DOIPubMedGoogle Scholar
- Vincent AL, Perez DR, Rajao D, Anderson TK, Abente EJ, Walia RR, et al. Influenza A virus vaccines for swine. Vet Microbiol. 2017;206:35–44. DOIPubMedGoogle Scholar
- Lewis NS, Anderson TK, Kitikoon P, Skepner E, Burke DF, Vincent AL. Substitutions near the hemagglutinin receptor-binding site determine the antigenic evolution of influenza A H3N2 viruses in U.S. swine. J Virol. 2014;88:4752–63. DOIPubMedGoogle Scholar
- Vincent A, Awada L, Brown I, Chen H, Claes F, Dauphin G, et al. Review of influenza A virus in swine worldwide: a call for increased surveillance and research. Zoonoses Public Health. 2014;61:4–17. DOIPubMedGoogle Scholar
- Viboud C, Bjørnstad ON, Smith DL, Simonsen L, Miller MA, Grenfell BT. Synchrony, waves, and spatial hierarchies in the spread of influenza. Science. 2006;312:447–51. DOIPubMedGoogle Scholar
- Lam TT-Y, Ip HS, Ghedin E, Wentworth DE, Halpin RA, Stockwell TB, et al. Migratory flyway and geographical distance are barriers to the gene flow of influenza virus among North American birds. Ecol Lett. 2012;15:24–33. DOIPubMedGoogle Scholar
- Petry M, Paarlberg PL, Lee JG. PRRS and the North American swine trade: a trade barrier analysis. Journal of Agricultural and Applied Economics. 1999;33:425–36. DOIGoogle Scholar
- Escalera-Zamudio M, Cobián-Güemes G, de los Dolores Soto-del Río M, Isa P, Sánchez-Betancourt I, Parissi-Crivelli A, et al. Characterization of an influenza A virus in Mexican swine that is related to the A/H1N1/2009 pandemic clade. Virology. 2012;433:176–82. DOIPubMedGoogle Scholar
1Current affiliation: Kansas State University, Manhattan, Kansas, USA.