Volume 25, Number 5—May 2019
Dispatch
Genetic Characterization of Middle East Respiratory Syndrome Coronavirus, South Korea, 2018
Figure 1
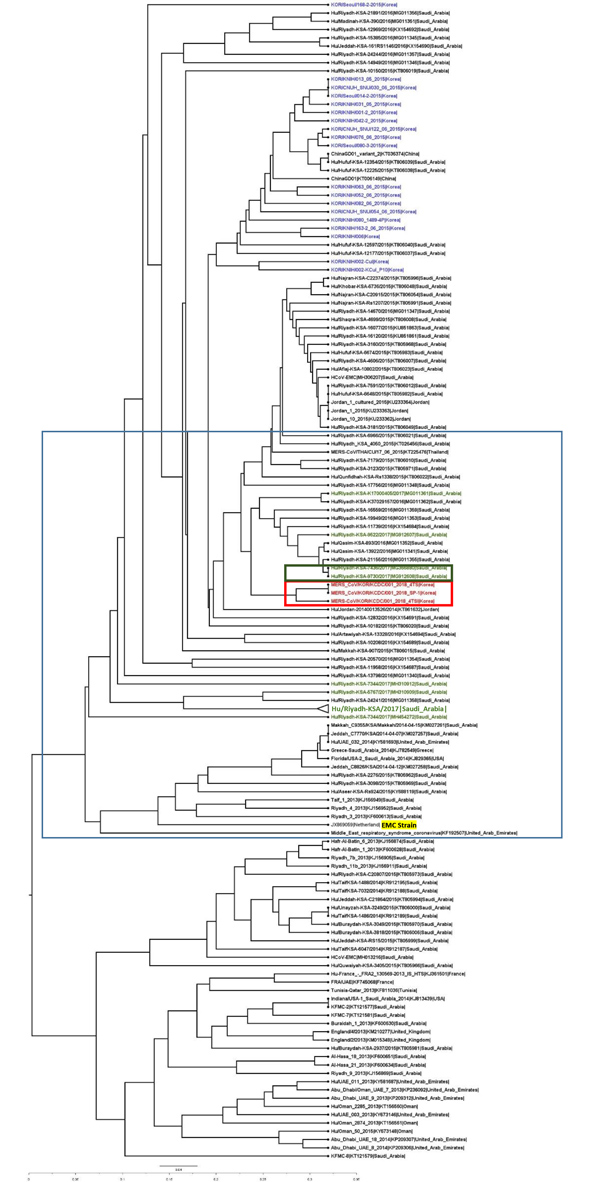
Figure 1. Molecular phylogenetic tree and coding region variants for the spike (S) glycoprotein gene of MERS-CoV isolates from South Korea, September 2018, and reference sequences. Phylogenetic analysis of 157 S sequences was performed using MEGA7 (https://www.megasoftware.net), with tree visualization using FigTree version 1.4.3 (http://tree.bio.ed.ac.uk/software/figtree). The taxonomic positions of circulating strains from the outbreak in South Korea and Riyadh, Saudi Arabia, are indicated. Boldface indicates compressed major clades of MERS-CoV. Bootstrap values (>70%) on nodes are shown as percentages based on 1,000 replicates. Red indicates South Korea 2018 isolates, blue indicates South Korea 2015 isolates, and green indicates Saudi Arabia 2017 isolates. The highlighted strain, from 2012, is the prototype strain used as the reference. EMC, Erasmus Medical Center; MERS-CoV, Middle East respiratory syndrome coronavirus. Scale bar indicates nucleotide substitutions per site.