Volume 25, Number 6—June 2019
Dispatch
Multirecombinant Enterovirus A71 Subgenogroup C1 Isolates Associated with Neurologic Disease, France, 2016–2017
Figure 2
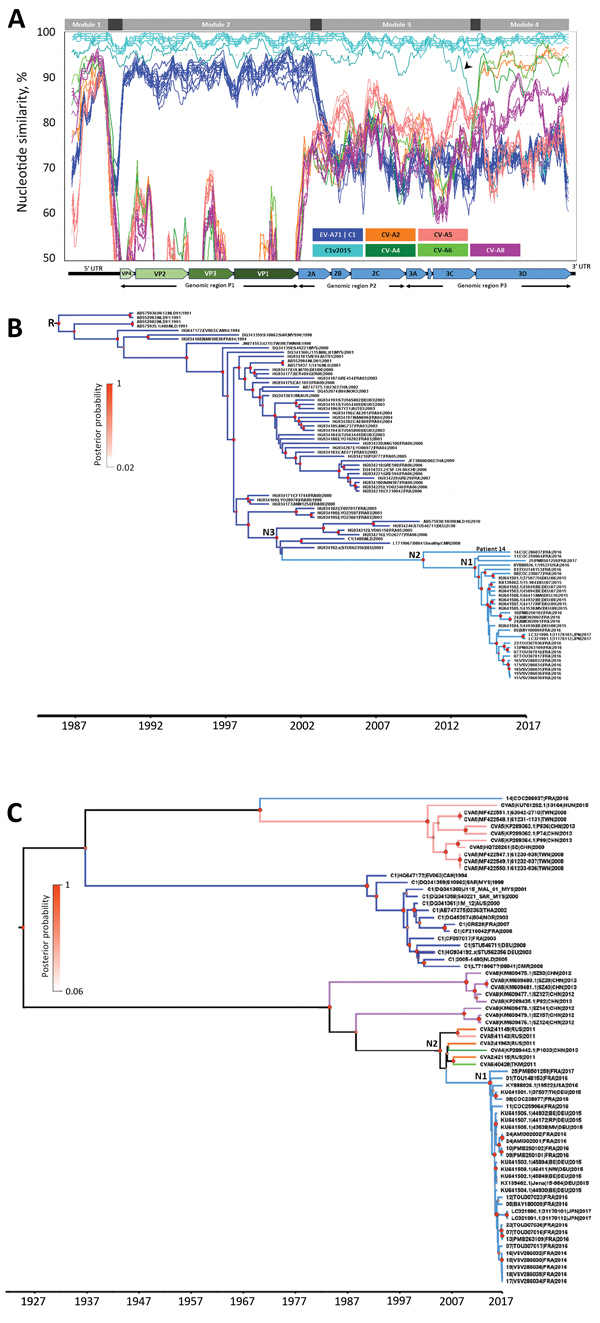
Figure 2. Nucleotide similarity and phylogenetic analyses of EV-A71 subgenogroup C1v2015 isolates, France, 2016–2017, constructed to determine temporal origin of C1v2015 lineage. A) Nucleotide similarity patterns between EV-A71 C1v2015 and other EV-A lineages indicate the C1v2015 genome has a mosaic structure. The genome of the virus from patient 10’s throat swab (10|PMB250102|FRA|2016) was used as the query sequence. The similarity plots determined with the other C1v2015 genomes (except 14|COC286037|FRA|2016) were similar. A schematic diagram of the enterovirus genome is shown at the bottom of the panel. Nucleotide similarity was calculated by the sliding window method (window of 300 nt moving every 30 nt). Four genomic modules (labeled at top of panel) with different genetic origins are identified. The 99% CIs of the nucleotide boundaries assessed for the genomic modules (indicated in dark gray) were determined as described in Hassel et al. (11). The 3′ end of module 1 and 5′ end of module 2 were located at the end of the 5′ UTR but were not determined precisely. The 3′ end of module 2 was located between nucleotides 3,532 and 3,722. The 5′ end of module 4 was located at the end of the 3Cpro gene (nucleotides 5,968–6,044). The arrowhead indicates a previously undescribed recombinant lineage of C1v2015 (Appendixhttps://wwwnc.cdc.gov/EID/article/25/6/18-1460-App1.pdf). B) Phylogenetic tree constructed by using genomic region P1, encoding capsid proteins VP1–VP4, and methods described earlier (11). We performed this analysis with 85 sequences assigned to the EV-A71 C1 and C1v2015 lineages. Tree shows the temporal distribution of lineages, including the emergence of lineage C1v2015. The sequences used as references were labeled with GenBank accession numbers. C) Phylogenetic tree constructed by using the 3Dpol gene encoding the viral RNA polymerase common to C1v2015 and several CV-A strains. The dataset comprised 70 sequences: 24 CV-A (including 5 sequences from this study), 14 EV-A71 C1 (including 6 sequences from this study), 12 publicly available C1v2015, and 20 C1v2015 sequences from this study. Recombination analyses provided no evidence of internal breakpoints within the sequences. N1 represents the time to most recent common ancestor (MRCA) of all included EV-A71 C1v2015 isolates except the virus from patient 14; N2 in panel B represents the MRCA of all EV-A71 C1v2015 isolates, including the virus from patient 14; N2 in panel C represents the MRCA of EV-A71 C1v2015 and its parent C1 lineage; and N3 represents the MRCA of EV-A71 C1v2015 and its parent C1 lineage. Diameters of circles at nodes reflect posterior probability. Branches of trees are color coded according to virus lineage as indicated in panel A. AUS, Australia; AUT, Austria; CAN, Canada; CHE, Switzerland; CHN, China; CMR, Cameroon; C1v2015, enterovirus subgenogroup C1 strain discovered in 2015; CV-A, coxsackievirus A; DEU, Germany; EV-A71C1, enterovirus A71 subgenogroup C1; FRA, France; JPN, Japan; N, node; NLD, the Netherlands; NOR, Norway; MYS, Malaysia; RUS, Russia; THA, Thailand; TKM, Turkmenistan; TWN, Taiwan; USA, United States; UTR, untranslated region; VP, viral protein.
References
- Bessaud M, Razafindratsimandresy R, Nougairède A, Joffret ML, Deshpande JM, Dubot-Pérès A, et al. Molecular comparison and evolutionary analyses of VP1 nucleotide sequences of new African human enterovirus 71 isolates reveal a wide genetic diversity. PLoS One. 2014;9:
e90624 . DOIPubMedGoogle Scholar - Hassel C, Mirand A, Lukashev A, TerletskaiaLadwig E, Farkas A, Schuffenecker I, et al. Transmission patterns of human enterovirus 71 to, from and among European countries, 2003 to 2013. Euro Surveill. 2015;20:30005. DOIPubMedGoogle Scholar
- Antona D, Kossorotoff M, Schuffenecker I, Mirand A, Leruez-Ville M, Bassi C, et al. Severe paediatric conditions linked with EV-A71 and EV-D68, France, May to October 2016. Euro Surveill. 2016;21:30402. DOIPubMedGoogle Scholar
- Böttcher S, Obermeier PE, Neubauer K, Diedrich S; Laboratory Network for Enterovirus Diagnostics. Recombinant enterovirus A71 subgenogroup C1 strains, Germany, 2015. Emerg Infect Dis. 2016;22:1843–6. DOIPubMedGoogle Scholar
- Karrasch M, Fischer E, Scholten M, Sauerbrei A, Henke A, Renz DM, et al. A severe pediatric infection with a novel enterovirus A71 strain, Thuringia, Germany. J Clin Virol. 2016;84:90–5. DOIPubMedGoogle Scholar
- Casas-Alba D, de Sevilla MF, Valero-Rello A, Fortuny C, García-García JJ, Ortez C, et al. Outbreak of brainstem encephalitis associated with enterovirus-A71 in Catalonia, Spain (2016): a clinical observational study in a children’s reference centre in Catalonia. Clin Microbiol Infect. 2017;23:874–81. DOIPubMedGoogle Scholar
- Wieczorek M, Purzyńska M, Krzysztoszek A, Ciąćka A, Figas A, Szenborn L. Genetic characterization of enterovirus A71 isolates from severe neurological cases in Poland. J Med Virol. 2018;90:372–6. DOIPubMedGoogle Scholar
- Midgley SE, Nielsen AG, Trebbien R, Poulsen MW, Andersen PH, Fischer TK. Co-circulation of multiple subtypes of enterovirus A71 (EV- A71) genotype C, including novel recombinants characterised by use of whole genome sequencing (WGS), Denmark 2016. Euro Surveill. 2017;22:30565. DOIPubMedGoogle Scholar
- Lukashev AN, Shumilina EY, Belalov IS, Ivanova OE, Eremeeva TP, Reznik VI, et al. Recombination strategies and evolutionary dynamics of the Human enterovirus A global gene pool. J Gen Virol. 2014;95:868–73. DOIPubMedGoogle Scholar
- van der Sanden S, van der Avoort H, Lemey P, Uslu G, Koopmans M. Evolutionary trajectory of the VP1 gene of human enterovirus 71 genogroup B and C viruses. J Gen Virol. 2010;91:1949–58. DOIPubMedGoogle Scholar
- Hassel C, Mirand A, Farkas A, Diedrich S, Huemer HP, Peigue-Lafeuille H, et al.; HFMD French Study Network. Phylogeography of coxsackievirus A16 reveals global transmission pathways and recent emergence and spread of a recombinant genogroup. J Virol. 2017;91:
e00630-17 . DOIPubMedGoogle Scholar - Chumakov M, Voroshilova M, Shindarov L, Lavrova I, Gracheva L, Koroleva G, et al. Enterovirus 71 isolated from cases of epidemic poliomyelitis-like disease in Bulgaria. Arch Virol. 1979;60:329–40. DOIPubMedGoogle Scholar
- Nagy G, Takátsy S, Kukán E, Mihály I, Dömök I. Virological diagnosis of enterovirus type 71 infections: experiences gained during an epidemic of acute CNS diseases in Hungary in 1978. Arch Virol. 1982;71:217–27. DOIPubMedGoogle Scholar
- McWilliam Leitch EC, Cabrerizo M, Cardosa J, Harvala H, Ivanova OE, Koike S, et al. The association of recombination events in the founding and emergence of subgenogroup evolutionary lineages of human enterovirus 71. J Virol. 2012;86:2676–85. DOIPubMedGoogle Scholar
- Kyriakopoulou Z, Pliaka V, Amoutzias GD, Markoulatos P. Recombination among human non-polio enteroviruses: implications for epidemiology and evolution. Virus Genes. 2015;50:177–88. DOIPubMedGoogle Scholar