Volume 26, Number 12—December 2020
Dispatch
Differential Tropism of SARS-CoV and SARS-CoV-2 in Bat Cells
Figure 2
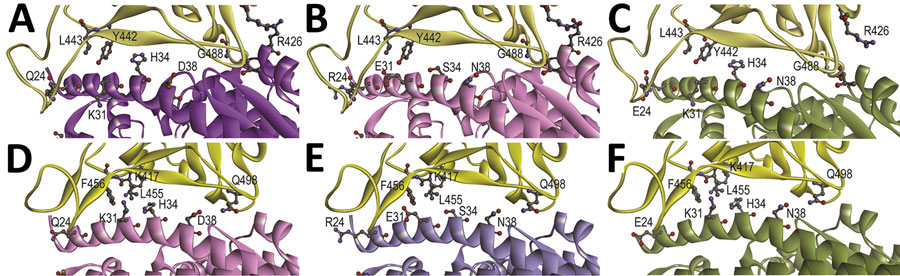
Figure 2. Structural modeling of the human (A, D), Rhinolophus sinicus bat (Rs-bat) (B, E), and Pipistrellus abramus bat (Pa-bat) (C, F) ACE2 with the receptor-binding domain (RBD) of the spike proteins of SARS-CoV and SARS-CoV-2. The models of RBDs of SARS-CoV and SARS-CoV-2 (yellow) are shown with human (purple), Rs-bat (pink). and Pa-bat (green) ACE2 structures in ribbon diagrams. The interface of different RBDs and human/bat ACE2 are shown and the residues with potential impact on binding affinity are shown in ball-and-stick format. Images were produced using Discovery Studio visualizer (Accelrys, https://www.accelrys.com).
1These authors contributed equally to this article.
Page created: July 17, 2020
Page updated: November 19, 2020
Page reviewed: November 19, 2020
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.