Volume 26, Number 2—February 2020
Research
Unique Clindamycin-Resistant Clostridioides difficile Strain Related to Fluoroquinolone-Resistant Epidemic BI/RT027 Strain
Cite This Article
Citation for Media
Abstract
During a surveillance study of patients in a long-term care facility and the affiliated acute care hospital in the United States, we identified a Clostridioides difficile strain related to the epidemic PCR ribotype (RT) 027 strain associated with hospital outbreaks of severe disease. Fifteen patients were infected with this strain, characterized as restriction endonuclease analysis group DQ and RT591. Like RT027, DQ/RT591 contained genes for toxin B and binary toxin CDT and a tcdC gene of identical sequence. Whole-genome sequencing and multilocus sequence typing showed that DQ/RT591 is a member of the same multilocus sequence typing clade 2 as RT027 but in a separate cluster. DQ/RT591 produced a similar cytopathic effect as RT027 but showed delayed toxin production in vitro. DQ/RT591 was susceptible to moxifloxacin but highly resistant to clindamycin. Continued surveillance is warranted for this clindamycin-resistant strain that is related to the fluoroquinolone-resistant epidemic RT027 strain.
As the leading cause of healthcare-associated infectious diarrhea and colitis, Clostridioides (formerly Clostridium) difficile continues to affect patients in hospitals and extended-care facilities in the United States (1–3). Among the numerous C. difficile strains, none have been more important to the healthcare community at large than the strain characterized as toxinotype III, restriction endonuclease analysis (REA) group BI, PCR ribotype (RT) 027, and sequence type (ST) 1, also known as pulsed-field gel electrophoresis type NAP1 (4,5). Because the BI/RT027 strain has been associated with numerous healthcare facility outbreaks and an increasing number of illnesses and deaths, it has been called hypervirulent (6). Factors that potentially contribute to increased virulence for this strain include increased sporulation, particular DNA gyrase mutations, the ability to survive in hostile environments, and increased toxin production (7–9).
The pathogenicity locus of BI/RT027 includes a characteristic 18-bp deletion and a single-base deletion at position 117 of the tcdC gene (4). These mutations result in a truncated TcdC protein that is rendered nonfunctional, leading to a lack of regulation of the tcdA and tcdB genes and potentially increased toxin A and B production (4). Virulence may be further affected by the binary toxin (CDT), which is coded for by genes located outside the pathogenicity locus, and its proposed role is to increase adherence of the bacterium to the epithelium (4,10).
Presumptive detection of the BI/RT027 strain by the Xpert C. difficile EPI assay (Cepheid, https://www.cepheid.com) is based on PCR amplification of targets, including the deletion at position 117 within tcdC and sequences within tcdB and the binary CDT genes (11). During the past 10 years, non-RT027 strains have emerged that test positive by this assay because they have the same or similar gene targets (12,13). Several of these non-RT027 strains also have been associated with increased numbers of illnesses and deaths, suggesting that the gene targets for this assay may be related to increased virulence (12–14).
In 2012, during a surveillance study at 2 US Veteran Affairs (VA) long-term care facilities (LTCFs) and their affiliated acute care facilities, we detected a clonal C. difficile outbreak at 1 of the LTCFs and the affiliated acute care facility (15). The organism was initially identified as the epidemic strain BI/RT027 after it tested positive by Xpert C. difficile Epi assay (15). However, further analysis identified this strain as REA group DQ and RT591. We report the microbiological and molecular characterization of this strain and the clinical findings of the infected patients.
During February 2012–August 2012, we obtained swab specimens from the perirectal area of asymptomatic LTCF patients at 2 VA facilities in Cleveland, Ohio, and Chicago, Illinois, USA, as part of a validation study comparing PCR using the Xpert C. difficile Epi assay and culture (15). In addition, fecal specimens from symptomatic patients with C. difficile infection (CDI) were cultured for C. difficile as part of a larger surveillance study of C. difficile in each LTCF and the associated acute care facility at these 2 sites (16). Specimens obtained from the perirectal area using BD BBL CultureSwabs (Becton Dickinson, https://www.bd.com) and submitted fecal specimens from symptomatic patients were cultured anaerobically on selective media as previously described (15). We reviewed medical records to obtain information about demographics, medical conditions, medications, CDI treatment, and outcomes. The severity of CDI and determination of initial versus recurrent cases were classified in accordance with the Infectious Diseases Society of America and Society for Healthcare Epidemiology of America C. difficile infection guidelines (17). The institutional review board of each hospital approved the study protocol.
REA
Recovered C. difficile isolates were first subjected to typing by REA. Using the methods provided by Clabots et al. (18), total cellular DNA was purified and subjected to HindIII restriction enzyme digestion and electrophoresis. The resulting banding patterns were then compared with a known database. All isolates representing a newly recognized REA type that corresponded to a presumptive BI/RT027 identified by the Xpert C. difficile Epi assay were subjected to PCR ribotyping, whole-genome sequencing (WGS), multilocus sequence typing (MLST), and PCR amplification of cdtA, cdtB, tcdA, and tcdB and an 18-bp deletion in tcdC. Sequencing of tcdC (1 isolate), toxinotyping (1 isolate), toxin production in vitro (1 isolate per REA strain group), and antimicrobial susceptibility testing (6 isolates) were performed on selected isolates.
PCR Ribotyping
We characterized recovered C. difficile isolates using high-resolution capillary gel electrophoresis–based PCR ribotyping. We analyzed these isolates against a library of standard profiles, as described previously in the internationally validated consensus protocol from Fawley et al. (19).
Gene Analysis of Binary Toxin CDT, Toxins A and B, and the Negative Toxin Regulator
We conducted PCR amplification of cdtA, cdtB, tcdA, and tcdB and an 18-bp deletion in tcdC on all isolates identified as BI/RT027 and DQ/RT591 as previously described (20). In addition, by using primers previously described by Rupnik et al. (21), we amplified cdtB and tcdB by PCR to confirm the presence of CDT and toxin B on a representative DQ/RT591 isolate followed by amplification and sequencing of the tcdC gene. A full-length tcdC PCR was performed using the following primers to produce a 910-bp product: forward primer ACTGTTTATTTGCAATTATAAAAACATCT; reverse primer TTACTTTATTTTGTAAAATTATGCTTAGGG. PCR amplicons were gel purified, sequenced, and compared with BI and strain 630.
Toxinotyping
We conducted toxinotyping on a representative DQ/RT591 isolate by performing restriction fragment-length polymorphism PCR of the B1 and A3 fragment. We assessed for variation in the first 3-kbp of tcdB and a repetitive 3-kbp fragment in tcdA (21).
WGS and MLST
We conducted WGS using a Nextera kit (Illumina, https://www.illumina.com) to prepare genomic DNA libraries and sequenced the RT591 organism using an Illumina MiSeq producing 2×250-bp read sets in accordance with the manufacturer’s protocols. Reads were filtered with SolexaQA++ 3.1 by dynamic trimming bases lower than Phred 30 and discarding reads <50-bp long. (22) We then assembled high-fidelity filtered reads into contigs at least 500 bp long with SPAdes 3.6.2 (22,23). Isolates that met the following 5 molecular testing criteria (positive for cdtA, cdtB, tcdA, and tcdB and an 18-bp deletion in tcdC) were then analyzed by genomewide average nucleotide identity (ANI). We computed pairwise ANI values between genomes from NUCmer v3.1 alignments aided by PyANI version 0.1 (24). Pairwise identity values were sorted into a 2-dimensional matrix with pandas version 0.15.2 in Python, and a heatmap of identity values with hierarchical clustering linkages was visualized with gplots version 2.16.0 in R (25). DQ/RT591 genomes also had single-nucleotide polymorphisms (SNPs) identified with Parsnp version 1.3.
We retrieved all STs on PubMLST’s webserver (https://pubmlst.org/cdifficile) (351 as of June 28, 2016) to compare distances among STs. The database of profiles was based on the first scheme created for C. difficile, which uses the loci of adk, atpA, dxr, glyA, recA, sodA, and tpi. Of the 3,501-bp length for each ST, 524 positions were found to have a nucleotide variant among the 351 STs. The variable sites were given to RAxML version 8.1.17 with the generalized time reversible substitution model, and the tree was visualized in Figtree version 1.4.2 (26).
In Vitro Toxin Production and Antimicrobial Susceptibility Testing
We determined toxin production on C. difficile isolate supernatants of representative isolates of 5 different REA group strains after 24, 48, and 72 hours of growth in brain heart infusion broth media (27). Toxin concentrations were determined by enzyme immunoassay (C. difficile toxA/B II EIA; TechLab, https://www.techlab.com) and interpolation from a standard curve using a toxin A standard of known concentration. Assays were performed in triplicate on a representative DQ isolate and compared with supernatants from toxigenic strains BI (RT027), J (RT001), AF (RT244), and a nontoxigenic strain, REA group T. A qualitative cytotoxin analysis was performed on the representative isolate supernatants using human fibroblast cells (Bartels cytotoxicity assay; Trinity Biotech, https://trinitylifesciences.com). We determined antimicrobial susceptibilities by Etest (bioMérieux, https://www.biomerieux-usa.com) for moxifloxacin, ceftriaxone, azithromycin, and clindamycin on taurocholate fructose agar plates (28,29). We confirmed the susceptibility results for moxifloxacin and clindamycin by testing 5 additional unique strains of BI and DQ recovered from patients at the VA site where the DQ outbreak occurred. We used Clinical and Laboratory Standards Institute guidelines for resistance cutoff values (30,31).
Statistical Analysis
We compared characteristics and outcomes of patients colonized or infected with BI/NAP1/027 and other strain types with those of patients colonized or infected with DQ/591 strains. Student t-test was used for normally distributed data and Fisher exact test for categorical data. We analyzed data using SPSS Statistics 10.0 (IBM, https://www.ibm.com).
REA group BI strains were the most common strains recovered from both VA sites and accounted for 33 (40%) of the 83 isolates at the Cleveland site. In addition, 16 (19%) of the isolates from 15 Cleveland patients were identified as REA strain DQ, even though the corresponding Xpert C. difficile Epi assay results indicated the presence of the NAP1 strain (i.e., REA group BI). No DQ strains were found at the Chicago site.
We compared baseline characteristics and outcomes of the 15 patients with fecal cultures positive for the DQ strain with those of the 22 patients with BI/NAP1/027 strains and 27 with other strain types (Table). Ten (67%) of the 15 patients with the DQ strain were LTCF residents, 4 (27%) were on the spinal cord injury unit, and 1 was hospitalized on a medical ward. Of the 7 patients with CDI caused by DQ strains, 3 (43%) met criteria for severe CDI, but none had fulminant CDI. All 7 CDI cases were healthcare associated; 3 of these patients had onset in the hospital, and 4 had onset in the LTCF. In all patients with CDI caused by DQ strains, diarrhea resolved with therapy, but CDI recurred in 3. Patients colonized or infected with DQ strains were significantly more likely than those with BI/NAP1/027 or other strain types to be LTCF residents and to have received antimicrobial drugs during the past 90 days. Patients with other strain types were significantly less likely than patients with DQ strains to have a recent intensive care unit admission, to have healthcare-associated CDI, or to die within 6 months after the CDI diagnosis.
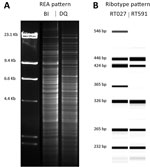
The HindIII REA banding patterns differed between the DQ and BI strains (Figure 1, panel A). The PCR ribotype patterns were more similar, except for the RT591 pattern (DQ), which was missing 2 major bands present in RT027 pattern (BI) (Figure 1, panel B).
PCR amplification of tcdB and cdtB in a representative DQ isolate indicated the presence of genes for toxin B and binary toxin CDT, consistent with the profile of the epidemic BI/RT027 strain (data not shown). In addition, amplification and sequencing of the tcdC gene showed complete alignment with tcdC from a reference BI strain, both of which contained an 18-bp deletion from positions 316 to 333 and a single base deletion at position 117 that resulted in a stop codon at position 196, unlike the reference C. difficile strain, 630 (data not shown) (32). The similarities to the BI/RT027 strain were further validated because toxinotyping indicated the DQ/591 strain was toxinotype III (data not shown).
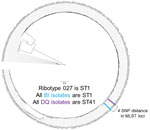
WGS and MLST results showed that, although the strains are closely related and reside within the same clade (MLST clade 2), they form a separate cluster (Figure 2). ANI showed a clear separation of the BI/027 and DQ/591 isolates at the whole genome level (Appendix Figure 1). By MLST analysis, the DQ/RT591 isolates were ST41, whereas BI/RT027 were ST1. Although the STs are closely related, there is a 4 SNP separation in the MLST loci.
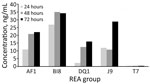
Supernatant from the DQ/RT591 strain produced typical cellular rounding on fibroblasts similar to BI/RT027, whereas the AF/RT244 strain produced a different phenotype, clumping and rounding of cells (Appendix Figure 2). Strain AF/RT244 is also related to RT027 and was responsible for an outbreak of severe disease in Australia (12). DQ/RT591 showed minimal toxin production in vitro at 24 hours; by 48 hours, toxin levels were similar to those of strain J/RT001 but still less than levels produced by BI/RT027 (Figure 3).
The DQ/RT591 strain was highly resistant to clindamycin (MIC >256 μg/mL) and azithromycin (MIC >256 μg/mL) but was susceptible to moxifloxacin (MIC range 1–2 μg/mL) and ceftriaxone (MIC 16 μg/mL). BI/RT027 was resistant to moxifloxacin (MIC >32 μg/mL) and azithromycin (MIC >256 μg/mL) and variably resistant to clindamycin (MIC range 6–32 μg/mL). Furthermore, WGS revealed that all DQ/RT591 isolates contained a variant ermB sequence known to confer clindamycin resistance (33).
During a surveillance study of C. difficile in asymptomatic LTCF patients and symptomatic patients in the affiliated acute care hospitals at 2 VA facilities (16), we detected a clonal outbreak of a newly recognized C. difficile strain at the Cleveland facility. This strain, identified as REA group DQ, ribotype 591, is closely related to the BI/RT027 strain, and a commercial PCR erroneously identified it as the epidemic NAP1 strain (i.e., BI/RT027). The strain was misidentified as NAP1 by Xpert C. difficile PCR (Cepheid) because of the similar genetic findings within the C. difficile pathogenicity locus, and the presence of the binary toxin CDT gene cdtB (33). Our findings and previous reports of other non-RT027 strains show that these genetic targets are not specific to NAP1 (12,13). Even though the outbreak of RT244 in Melbourne, Australia, was misidentified as NAP1 (RT027) by the same commercial PCR, it was associated with severe disease in the infected patients (12), suggesting the possibility of shared virulence determinants.
DQ/RT591 and BI/RT027 share several characteristics, including tcdB, cdtB, the 18-bp and position 117 tcdC deletions, and a similar cytotoxic phenotype. Despite these shared genetic and phenotypic characteristics, in vitro toxin production appeared to be delayed and somewhat lower in the DQ/RT591 strain than in BI/RT027. The clinical manifestations of the patients colonized or infected with DQ/RT591 were not unusual and did not differ substantially from those of patients with BI/NAP1/027 and other strain types in the cohort reported here. In nearly half (47%) of the patients, symptoms developed that were consistent with CDI. Although no fulminant CDI cases or deaths directly related to C. difficile were recognized, in CDI that developed because of DQ/591, 43% were classified as severe CDI in accordance with Infectious Diseases Society of America and Society for Healthcare Epidemiology of America guidelines. Likewise, no fulminant CDI cases or CDI-related deaths were recognized among the patients infected with BI/RT027 during this same study. However, all-cause mortality was lower for persons with non-DQ/RT591 and non-BI/RT027 infections. All the documented transmission events occurred in the BI/RT027 patients (34). Despite the fact that all the CDI cases caused by DQ/RT591 were healthcare associated, WGS did not identify any transmission events between patients because core genome SNP differences were >8 among DQ isolates (Appendix Figure 3) (34).
All patients in whom DQ/RT591 was confirmed had received antimicrobial drugs within 90 days before testing, and all patients with a CDI were classified as having a healthcare-associated infection; nearly all cases occurred in the LTCF or spinal cord injury unit. No specific antimicrobial drug was highly associated with infection by this strain. Fluoroquinolones have been associated with CDI outbreaks with the BI/RT027 strain, which is highly resistant to fluoroquinolones in vitro (35). Among the DQ/RT591- and BI/RT027-infected patients, receipt of fluoroquinolones or clindamycin was limited. Despite high-level resistance to clindamycin in vitro and the presence of ermB in DQ/RT591, only 1 patient infected with this strain had received clindamycin.
Our experience was limited, but DQ/RT591 did not appear to carry the same level of clinical severity that BI/RT027 has exhibited (6). This difference in severity might be attributable to delayed toxin production in vivo (Figure 3). Although increased toxin production has been proposed as the reason for increased virulence with the BI/RT027 strain (4,29), increased toxin production was not demonstrated for the AF/RT244 strain, which nevertheless was associated with an outbreak of increased severity in Australia (12). AF/RT244 also shows a different cytopathic effect than BI/027, which suggests the presence of a variant toxin B in AF/RT244 (4,12). Factors associated with increased virulence associated with these strains are still incompletely defined.
Because REA DQ/RT591 is closely related to BI/027, further monitoring is required to determine whether this strain carries risk for increased illness and death or has the capability of widespread dissemination. Since we completed this work, RT591 was reported as the most prevalent C. difficile strain in 3 tertiary hospitals in Colombia. These RT591 isolates were also mostly resistant to clindamycin (85%) (36).
Acknowledgments
The RT244 strain was a gift from T. Riley. We thank Adam Cheknis for technical support with the toxin and susceptibility assays. We also thank Brandi M. Limbago, Maria Karlsson, and Ashley L. Paulick for their support on this project.
This study was supported by the Centers for Disease Control and Prevention and by the Department of Veterans Affairs.
Dr. Skinner is an infectious diseases fellow at Loyola University Medical Center and the Hines Veterans Affairs Hospital. His research interests include C. difficile epidemiology, treatment, host response, and molecular characterization.
References
- Dallal RM, Harbrecht BG, Boujoukas AJ, Sirio CA, Farkas LM, Lee KK, et al. Fulminant Clostridium difficile: an underappreciated and increasing cause of death and complications. Ann Surg. 2002;235:363–72. DOIPubMedGoogle Scholar
- McDonald LC, Killgore GE, Thompson A, Owens RC Jr, Kazakova SV, Sambol SP, et al. An epidemic, toxin gene-variant strain of Clostridium difficile. N Engl J Med. 2005;353:2433–41. DOIPubMedGoogle Scholar
- Lessa FC, Mu Y, Bamberg WM, Beldavs ZG, Dumyati GK, Dunn JR, et al. Burden of Clostridium difficile infection in the United States. N Engl J Med. 2015;372:825–34. DOIPubMedGoogle Scholar
- O’Connor JR, Johnson S, Gerding DN. Clostridium difficile infection caused by the epidemic BI/NAP1/027 strain. Gastroenterology. 2009;136:1913–24. DOIPubMedGoogle Scholar
- Brazier JS, Raybould R, Patel B, Duckworth G, Pearson A, Charlett A, et al.; HPA Regional Microbiology Network. Distribution and antimicrobial susceptibility patterns of Clostridium difficile PCR ribotypes in English hospitals, 2007-08. Euro Surveill. 2008;13:19000. DOIPubMedGoogle Scholar
- See I, Mu Y, Cohen J, Beldavs ZG, Winston LG, Dumyati G, et al. NAP1 strain type predicts outcomes from Clostridium difficile infection. Clin Infect Dis. 2014;58:1394–400. DOIPubMedGoogle Scholar
- Merrigan M, Venugopal A, Mallozzi M, Roxas B, Viswanathan VK, Johnson S, et al. Human hypervirulent Clostridium difficile strains exhibit increased sporulation as well as robust toxin production. J Bacteriol. 2010;192:4904–11. DOIPubMedGoogle Scholar
- He M, Miyajima F, Roberts P, Ellison L, Pickard DJ, Martin MJ, et al. Emergence and global spread of epidemic healthcare-associated Clostridium difficile. Nat Genet. 2013;45:109–13. DOIPubMedGoogle Scholar
- Jump RLP, Pultz MJ, Donskey CJ. Vegetative Clostridium difficile survives in room air on moist surfaces and in gastric contents with reduced acidity: a potential mechanism to explain the association between proton pump inhibitors and C. difficile-associated diarrhea? Antimicrob Agents Chemother. 2007;51:2883–7. DOIPubMedGoogle Scholar
- Gerding DN, Johnson S, Rupnik M, Aktories K. Clostridium difficile binary toxin CDT: mechanism, epidemiology, and potential clinical importance. Gut Microbes. 2014;5:15–27. DOIPubMedGoogle Scholar
- Scardina T, Labuszewski L, Pacheco SM, Adams W, Schreckenberger P, Johnson S. Clostridium difficile infection (CDI) severity and outcome among patients infected with the NAP1/BI/027 strain in a non-epidemic setting. Infect Control Hosp Epidemiol. 2015;36:280–6. DOIPubMedGoogle Scholar
- Lim SK, Stuart RL, Mackin KE, Carter GP, Kotsanas D, Francis MJ, et al. Emergence of a ribotype 244 strain of Clostridium difficile associated with severe disease and related to the epidemic ribotype 027 strain. Clin Infect Dis. 2014;58:1723–30. DOIPubMedGoogle Scholar
- Kok J, Wang Q, Thomas LC, Gilbert GL. Presumptive identification of Clostridium difficile strain 027/NAP1/BI on Cepheid Xpert: interpret with caution. J Clin Microbiol. 2011;49:3719–21. DOIPubMedGoogle Scholar
- Wehrhahn MC, Keighley C, Kurtovic J, Knight DR, Hong S, Hutton ML, et al. A series of three cases of severe Clostridium difficile infection in Australia associated with a binary toxin producing clade 2 ribotype 251 strain. Anaerobe. 2019;55:117–23. DOIPubMedGoogle Scholar
- Donskey CJ, Sunkesula VC, Jencson AL, Stone ND, Gould CV, McDonald LC, et al. Utility of a commercial PCR assay and a clinical prediction rule for detection of toxigenic Clostridium difficile in asymptomatic carriers. J Clin Microbiol. 2014;52:315–8. DOIPubMedGoogle Scholar
- Pacheco SM, Donskey CJ, Samore M, Mayer J, Stone ND, Gould CV, et al. Clostridium difficile strains colonizing long-term care facility (LTCF) residents are similar to strains causing infection in both LTCF and hospital patients suggesting a shared continuum of transmission. Open Forum Infect Dis. 2014;1(Suppl 1):S437–8. DOIGoogle Scholar
- McDonald LC, Gerding DN, Johnson S, Bakken JS, Carroll KC, Coffin SE, et al. Clinical practice guidelines for Clostridium difficile infection in adults and children: 2017 update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA). Clin Infect Dis. 2018;66:987–94. DOIPubMedGoogle Scholar
- Clabots CR, Johnson S, Bettin KM, Mathie PA, Mulligan ME, Schaberg DR, et al. Development of a rapid and efficient restriction endonuclease analysis typing system for Clostridium difficile and correlation with other typing systems. J Clin Microbiol. 1993;31:1870–5.PubMedGoogle Scholar
- Fawley WN, Knetsch CW, MacCannell DR, Harmanus C, Du T, Mulvey MR, et al. Development and validation of an internationally-standardized, high-resolution capillary gel-based electrophoresis PCR-ribotyping protocol for Clostridium difficile. PLoS One. 2015;10:
e0118150 . DOIPubMedGoogle Scholar - Abrahamian FM, Talan DA, Krishnadasan A, Citron DM, Paulick AL, Anderson LJ, et al.; EMERGEncy ID NET Study Group. EMERGEncy ID NET Study Group. EMERGEncy ID NET Study Group. EMERGEncy ID NET Study Group. Clostridium difficile infection among US emergency department patients with diarrhea and no vomiting. Ann Emerg Med. 2017;70:19–27.e4. DOIPubMedGoogle Scholar
- Rupnik M, Brazier JS, Duerden BI, Grabnar M, Stubbs SLJ. Comparison of toxinotyping and PCR ribotyping of Clostridium difficile strains and description of novel toxinotypes. Microbiology. 2001;147:439–47. DOIPubMedGoogle Scholar
- Cox MP, Peterson DA, Biggs PJ, Solexa QA, Solexa QA. SolexaQA: At-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinformatics. 2010;11:485. DOIPubMedGoogle Scholar
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–77. DOIPubMedGoogle Scholar
- Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, et al. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. DOIPubMedGoogle Scholar
- Warnes GR, Bolker B, Bonebakker L, Gentleman R, Liaw WHA, Lumley T, et al. gplots: Various R programming tools for plotting data. R package version 2.16.0. 2016 [cited 2018 Nov 18]. https://CRAN.R-project.org/package=gplots
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–3. DOIPubMedGoogle Scholar
- Warny M, Pepin J, Fang A, Killgore G, Thompson A, Brazier J, et al. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet. 2005;366:1079–84. DOIPubMedGoogle Scholar
- Sambol SP, Tang JK, Merrigan MM, Johnson S, Gerding DN. Infection of hamsters with epidemiologically important strains of Clostridium difficile. J Infect Dis. 2001;183:1760–6. DOIPubMedGoogle Scholar
- Razaq N, Sambol S, Nagaro K, Zukowski W, Cheknis A, Johnson S, et al. Infection of hamsters with historical and epidemic BI types of Clostridium difficile. J Infect Dis. 2007;196:1813–9. DOIPubMedGoogle Scholar
- Clinical and Laboratory Standards Institute. Methods for antimicrobial susceptibility testing of anaerobic bacteria; approved standard: eighth edition (M11-A8). Wayne (PA): The Institute; 2012.
- Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; twenty-seventh edition (M100-S27). Wayne (PA): The Institute; 2017.
- Curry SR, Marsh JW, Muto CA, O’Leary MM, Pasculle AW, Harrison LH. tcdC genotypes associated with severe TcdC truncation in an epidemic clone and other strains of Clostridium difficile. J Clin Microbiol. 2007;45:215–21. DOIPubMedGoogle Scholar
- Gupta SK, Padmanabhan BR, Diene SM, Lopez-Rojas R, Kempf M, Landraud L, et al. ARG-ANNOT (antibiotic resistance gene-ANNOTation), a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob Agents Chemother. 2014;58:212–20. DOIPubMedGoogle Scholar
- Donskey CJ, Sunkesula VCK, Stone ND, Gould CV, McDonald LC, Samore M, et al. Transmission of Clostridium difficile from asymptomatically colonized or infected long-term care facility residents. Infect Control Hosp Epidemiol. 2018;39:909–16. DOIPubMedGoogle Scholar
- Wieczorkiewicz JT, Lopansri BK, Cheknis A, Osmolski JR, Hecht DW, Gerding DN, et al. Fluoroquinolone and macrolide exposure predict Clostridium difficile infection (CDI) with the highly fluoroquinolone- and macrolide-resistant epidemic C. difficile strain, BI/NAP1/027. Antimicrob Agents Chemother. 2015;60:418–23. DOIPubMedGoogle Scholar
- Salazar CL, Reyes C, Atehortua S, Sierra P, Correa MM, Paredes-Sabja D, et al. Molecular, microbiological and clinical characterization of Clostridium difficile isolates from tertiary care hospitals in Colombia. PLoS One. 2017;12:
e0184689 . DOIPubMedGoogle Scholar
Figures
Table
Cite This ArticleOriginal Publication Date: December 29, 2019
Table of Contents – Volume 26, Number 2—February 2020
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Stuart Johnson, Edward Hines, Jr. VA Hospital, Infectious Diseases, Research Service/151, 5000 S 5th Ave, Hines, IL 60141, USA; email:; Andrew M. Skinner, Loyola University Medical Center, 2160 S 1st Ave, Maywood, IL 60153, USA
Top