Volume 26, Number 3—March 2020
Synopsis
Mycobacterium tuberculosis Complex Lineage 3 as Causative Agent of Pulmonary Tuberculosis, Eastern Sudan1
Figure 1
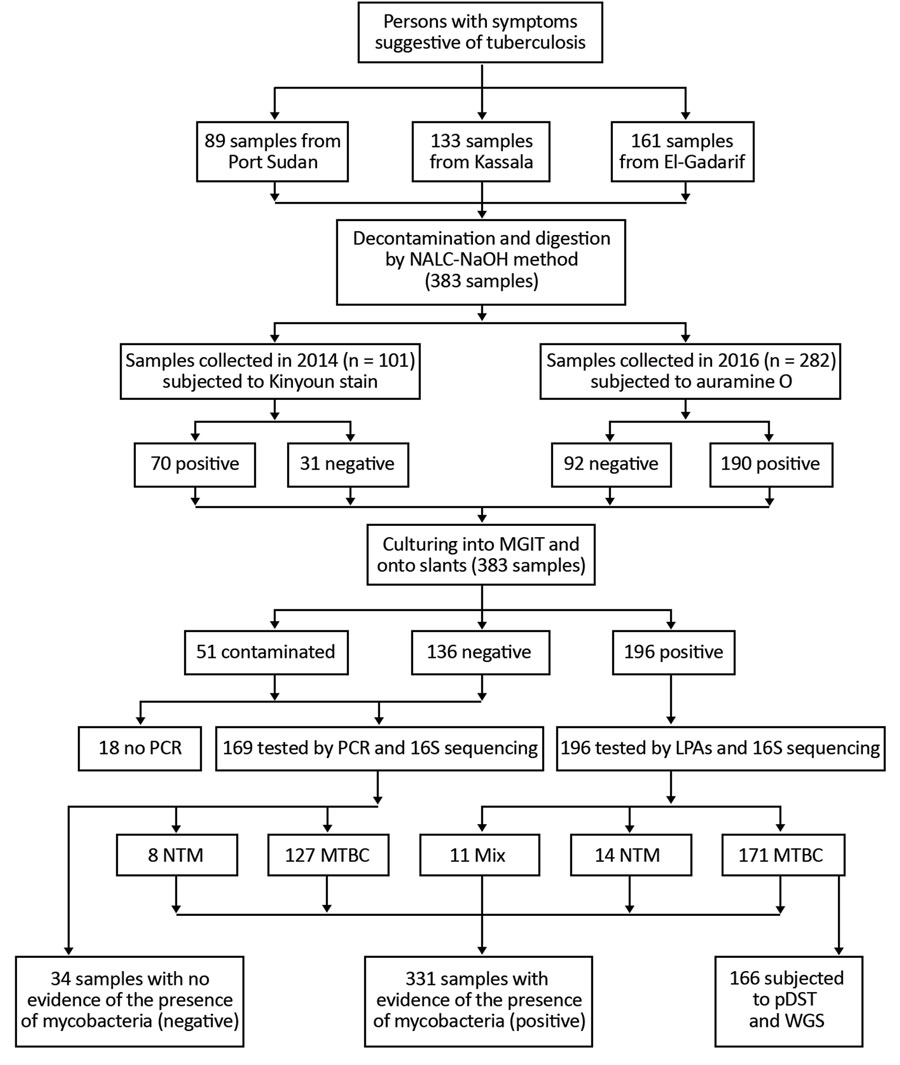
Figure 1. Work flow for study of Mycobacterium tuberculosis complex lineage 3 as causative agent of pulmonary tuberculosis, eastern Sudan. LPAs, HAIN line probe assay for GenoType CM and GenoType MTBC; MGIT, mycobacteria growth indicator tube; MTBC, Mycobacterium tuberculosis complex; mix, 2 different bacteria grew on the same culture; NALC-NaOH, sodium hydroxide/N-acetyl-cysteine; NTM, nontuberculous mycobacteria; pDST, phenotypic drug susceptibility testing; WGS, whole genome sequencing. Adopted from Shuaib et al. (14).
References
- World Health Organization. Global tuberculosis report. 2019 Sep 26 [cited 2019 Oct 30]. https://apps.who.int/iris/bitstream/handle/10665/329368/9789241565714-eng.pdf
- World Health Organization. Global tuberculosis report. 2012 Oct 17 [cited 2019 Mar 10]. https://www.who.int/tb/publications/global_report/gtbr12_main.pdf
- Abdallah TM, Ali AA. Epidemiology of tuberculosis in Eastern Sudan. Asian Pac J Trop Biomed. 2012;2:999–1001. DOIPubMedGoogle Scholar
- Muna OA. Determination of the prevalence of tuberculosis with drug-resistant strains of Mycobacterium tuberculosis in Khartoum, Gazira and camps for displaced people, Sudan. Oslo: Faculty of Medicine, University of Oslo; 2002.
- Sharaf Eldin GS, Fadl-Elmula I, Ali MS, Ali AB, Salih AL, Mallard K, et al. Tuberculosis in Sudan: a study of Mycobacterium tuberculosis strain genotype and susceptibility to anti-tuberculosis drugs. BMC Infect Dis. 2011;11:219. DOIPubMedGoogle Scholar
- Hassan SO, Musa MT, Elsheikh HM, Eleragi AM, Saeed OK. Drug resistance in Mycobacterium tuberculosis isolates from northeastern Sudan. Br J Med Res. 2012;2:424–33. DOIGoogle Scholar
- Abdul-Aziz AA, Elhassan MM, Abdulsalam SA, Mohammed EO, Hamid ME. Multi-drug resistance tuberculosis (MDR-TB) in Kassala state, eastern Sudan. Trop Doct. 2013;43:66–70. DOIPubMedGoogle Scholar
- Khalid FA, Hamid ZA, Mukhtar MM. Tuberculosis drug resistance isolates from pulmonary tuberculosis patients, Kassala State, Sudan. Int J Mycobacteriol. 2015;4:44–7. DOIPubMedGoogle Scholar
- Nour EMM, Saeed EMA, Zaki AZSA, Saeed NS. Drug resistance patterns of Mycobacterium tuberculosis isolates from patients with pulmonary tuberculosis in the Sudan. IOSR Journal of Dental and Medical Sciences. 2015;14:17–9.
- Eldirdery MM, Intisar EA, Mona OA, Fatima AK, Asrar MA, Nuha YI, et al. Prevalence of multidrug-resistant tuberculosis among smear positive pulmonary tuberculosis patients in eastern Sudan. Afr J Microbiol Res. 2017;5:32–6.
- Auld SC, Kasmar AG, Dowdy DW, Mathema B, Gandhi NR, Churchyard GJ, et al. Research roadmap for tuberculosis transmission science: where do we go from here and how will we know when we’re there? J Infect Dis. 2017;216(suppl_6):S662–8.
- Walker TM, Ip CL, Harrell RH, Evans JT, Kapatai G, Dedicoat MJ, et al. Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: a retrospective observational study. Lancet Infect Dis. 2013;13:137–46. DOIPubMedGoogle Scholar
- Merker M, Kohl TA, Niemann S, Supply P. The evolution of strain typing in the Mycobacterium tuberculosis complex. Adv Exp Med Biol. 2017;1019:43–78. DOIPubMedGoogle Scholar
- Shuaib YA, Khalil EAG, Schaible UE, Wieler LH, Bakheit MAM, Mohamed-Noor SE, et al. Smear microscopy for diagnosis of pulmonary tuberculosis in eastern Sudan. Tuberc Res Treat. 2018;2018:
8038137 . DOIPubMedGoogle Scholar - Khalid FA, Gasmelseed N, Hailu E, Eldirdery MM, Abebe M, Berg S, et al. Molecular identification of Mycobacterium tuberculosis causing Pulmonary Tuberculosis in Sudan. Eur Acad Res. 2016;4:7842–55.PubMedGoogle Scholar
- Gröschel MI, Walker TM, van der Werf TS, Lange C, Niemann S, Merker M. Pathogen-based precision medicine for drug-resistant tuberculosis. PLoS Pathog. 2018;14:
e1007297 . DOIPubMedGoogle Scholar - Meehan CJ, Goig GA, Kohl TA, Verboven L, Dippenaar A, Ezewudo M, et al. Whole genome sequencing of Mycobacterium tuberculosis: current standards and open issues. Nat Rev Microbiol. 2019;17:533–45. DOIPubMedGoogle Scholar
- Siddiqi S, Rüsch-Gerdes S. MGIT TM Procedure Manual. Geneva: Foundation for Innovative New Diagnostics (FIND); 2006.
- Deutsches Institut für Normung. Medical microbiology—diagnosis of tuberculosis. Part 3: detection of mycobacteria by culture methods. Berlin: The Institute; 2011.
- Somerville W, Thibert L, Schwartzman K, Behr MA. Extraction of Mycobacterium tuberculosis DNA: a question of containment. J Clin Microbiol. 2005;43:2996–7. DOIPubMedGoogle Scholar
- Hillemann D, Warren R, Kubica T, Rüsch-Gerdes S, Niemann S. Rapid detection of Mycobacterium tuberculosis Beijing genotype strains by real-time PCR. J Clin Microbiol. 2006;44:302–6. DOIPubMedGoogle Scholar
- Richter E, Niemann S, Gloeckner FO, Pfyffer GE, Rüsch-Gerdes S. Mycobacterium holsaticum sp. nov. Int J Syst Evol Microbiol. 2002;52:1991–6.PubMedGoogle Scholar
- Rüsch-Gerdes S, Pfyffer GE, Casal M, Chadwick M, Siddiqi S. Multicenter laboratory validation of the BACTEC MGIT 960 technique for testing susceptibilities of Mycobacterium tuberculosis to classical second-line drugs and newer antimicrobials. J Clin Microbiol. 2006;44:688–92. DOIPubMedGoogle Scholar
- Ängeby K, Juréen P, Kahlmeter G, Hoffner SE, Schön T. Challenging a dogma: antimicrobial susceptibility testing breakpoints for Mycobacterium tuberculosis. Bull World Health Organ. 2012;90:693–8. DOIPubMedGoogle Scholar
- Heyckendorf J, Andres S, Köser CU, Olaru ID, Schön T, Sturegård E, et al. What is resistance? Impact of phenotypic versus molecular drug resistance testing on therapy for multi- and extensively drug-resistant tuberculosis. Antimicrob Agents Chemother. 2018;62:e01550–17. PubMedGoogle Scholar
- Merker M, Barbier M, Cox H, Rasigade JP, Feuerriegel S, Kohl TA, et al. Compensatory evolution drives multidrug-resistant tuberculosis in Central Asia. eLife. 2018;7:
e38200 . DOIPubMedGoogle Scholar - Kohl TA, Utpatel C, Schleusener V, De Filippo MR, Beckert P, Cirillo DM, et al. MTBseq: a comprehensive pipeline for whole genome sequence analysis of Mycobacterium tuberculosis complex isolates. PeerJ. 2018;6:
e5895 . DOIPubMedGoogle Scholar - Price MN, Dehal PS, Arkin AP. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:
e9490 . DOIPubMedGoogle Scholar - He Z, Zhang H, Gao S, Lercher MJ, Chen WH, Hu S. Evolview v2: an online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 2016;44(W1):
W236-41 . DOIPubMedGoogle Scholar - Kannan L, Wheeler WC. Maximum parsimony on phylogenetic networks. Algorithms Mol Biol. 2012;7:9. DOIPubMedGoogle Scholar
- Feuerriegel S, Schleusener V, Beckert P, Kohl TA, Miotto P, Cirillo DM, et al. PhyResSE: a Web tool delineating Mycobacterium tuberculosis antibiotic resistance and lineage from whole-genome sequencing data. J Clin Microbiol. 2015;53:1908–14. DOIPubMedGoogle Scholar
- Kim HY. Statistical notes for clinical researchers: Chi-squared test and Fisher’s exact test. Restor Dent Endod. 2017;42:152–5. DOIPubMedGoogle Scholar
- Coll F, McNerney R, Guerra-Assunção JA, Glynn JR, Perdigão J, Viveiros M, et al. A robust SNP barcode for typing Mycobacterium tuberculosis complex strains. Nat Commun. 2014;5:4812. DOIPubMedGoogle Scholar
- Meehan CJ, Moris P, Kohl TA, Pečerska J, Akter S, Merker M, et al. The relationship between transmission time and clustering methods in Mycobacterium tuberculosis epidemiology. EBioMedicine. 2018;37:410–6. DOIPubMedGoogle Scholar
- Eldirdery MM, Alrayah IE, ElkareIm MOA, Khalid FA, Elegail AMA, Ibrahim NY, et al. Genotyping of pulmonary Mycobacterium tuberculosis isolates from Sudan using spoligotyping. Am J Microbiol Res. 2015;3:125–8.PubMedGoogle Scholar
- Couvin D, Reynaud Y, Rastogi N. Two tales: Worldwide distribution of Central Asian (CAS) versus ancestral East-African Indian (EAI) lineages of Mycobacterium tuberculosis underlines a remarkable cleavage for phylogeographical, epidemiological and demographical characteristics. PLoS One. 2019;14:
e0219706 . DOIPubMedGoogle Scholar - Stucki D, Brites D, Jeljeli L, Coscolla M, Liu Q, Trauner A, et al. Mycobacterium tuberculosis lineage 4 comprises globally distributed and geographically restricted sublineages. Nat Genet. 2016;48:1535–43. DOIPubMedGoogle Scholar
- Abu Shanab ME. Defaulting to anti-tuberculosis treatment: proportional and associated factors among internally displaced people around Khartoum state. Khartoum (Sudan): Faculty of Public and Environmental Health, University of Khartoum; 2003.
- Ali AO, Prins MH. Patient non adherence to tuberculosis treatment in Sudan: socio demographic factors influencing non adherence to tuberculosis therapy in Khartoum State. Pan Afr Med J. 2016;25:80. PubMedGoogle Scholar
- Giampaglia CM, Martins MC, Vieira GB, Vinhas SA, Telles MA, Palaci M, et al. Multicentre evaluation of an automated BACTEC 960 system for susceptibility testing of Mycobacterium tuberculosis. Int J Tuberc Lung Dis. 2007;11:986–91.PubMedGoogle Scholar
- CRyPTIC Consortium and the 100,000 Genomes Project. Prediction of susceptibility to first-line tuberculosis drugs by DNA sequencing. N Engl J Med. 2018;379:1403–15. DOIGoogle Scholar
- Andres S, Gröschel MI, Hillemann D, Merker M, Niemann S, Kranzer K. A diagnostic algorithm to investigate pyrazinamide and ethambutol resistance in rifampin-resistant Mycobacterium tuberculosis isolates in a low-incidence setting. Antimicrob Agents Chemother. 2019;63:e01798–18.PubMedGoogle Scholar
1Preliminary results from this study were presented at the 39th Annual Congress of the European Society of Mycobacteriology, July 1–4, 2018, Dresden, Germany.
2These authors contributed equally to this article.
Page created: February 20, 2020
Page updated: February 20, 2020
Page reviewed: February 20, 2020
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.