Volume 26, Number 5—May 2020
Synopsis
Biphasic Outbreak of Invasive Group A Streptococcus Disease in Eldercare Facility, New Zealand
Cite This Article
Citation for Media
Abstract
A 3-month outbreak of invasive group A Streptococcus disease at an eldercare facility, in which 5 persons died, was biphasic. Although targeted chemoprophylaxis contained the initial outbreak, a second phase of the outbreak occurred after infection control processes ended. To retrospectively investigate the genomic epidemiology of the biphasic outbreak, we used whole-genome sequencing and multiple bioinformatics approaches. Analysis of isolates from the outbreak and isolates prospectively collected during the outbreak response indicated a single S. pyogenes emm81 clone among residents and staff members. Outbreak isolates differed from nonoutbreak emm81 isolates by harboring an integrative conjugative genomic element that contained the macrolide resistance determinant erm(TR). This study shows how retrospective high-resolution genomic investigations identified rapid spread of a closed-facilty clonal outbreak that was controlled, but not readily cleared, by infection control management procedures.
Streptococcus pyogenes, or group A Streptococcus (GAS), is a gram-positive, human-adapted opportunistic bacterial pathogen. GAS causes a wide variety of clinical manifestations, from relatively benign self-limiting infections of the nasopharynx or skin to life-threatening invasive infections such as bacteremia, pneumonia, and necrotizing fasciitis (1). The incidence of invasive GAS infections is highest among older adults (2,3), particularly those living in long-term care facilities (2,4). Outbreaks of GAS infections are often linked with distinct epidemiologic markers such as emm type (5,6). emm typing is a sequence-based method that analyzes heterogeneity in the 5′ end of the ubiquitous emm gene that encodes the M-protein (7). Although emm typing provides useful information about the potential relatedness of outbreak isolates, whole-genome sequencing enables outbreak investigations to proceed with a far greater level of discrimination than single-gene typing of GAS isolates (8,9). For this study, we used multiple whole-genome–based approaches to examine the genetic relationships and molecular drivers of a biphasic GAS outbreak in an eldercare facility in which 14 persons became ill and 5 died.
Setting
During winter 2014, an outbreak of invasive and noninvasive GAS disease occurred in an eldercare facility in South Island, New Zealand. The outbreak was recognized by a senior laboratory scientist who noted the sudden increase in positive blood cultures from the facility. The outbreak occurred in 2 phases. The first phase started in late May 2014 and ended when the last case-patient (a resident) was hospitalized in early June 2014. The initial 3 case-patients were admitted within 24 hours of each other, and GAS was isolated from blood or tissue cultures from all 3 case-patients. During the first phase, 6 case-patients with confirmed GAS infection were hospitalized; 5 died of presumed sepsis. Outbreak investigations and control measures were subsequently implemented and included screening staff members and residents by collecting throat swab samples and providing targeted chemoprophylaxis for residents and staff members who were in direct contact with case-patients.
These interventions continued until early July 2014; however, in late July, the outbreak recurred and continued until mid August 2014. In this second phase, a staff member was hospitalized with severe pharyngitis, after which 5 residents were hospitalized with soft tissue infections, septicemia, or both.
Case Definitions
According to disk-diffusion testing, all GAS isolates collected during the first phase were susceptible to penicillin but resistant to erythromycin. Accordingly, a suspected case-patient was defined as any resident or staff member from the facility who was unwell from early June 2014 through mid-November 2014, and a confirmed case-patient was any person from the facility from whom erythromycin-resistant GAS was cultured from blood, throat, or skin samples. The definition of a suspected case-patient was kept intentionally broad because of the wide-ranging symptoms among initial case-patients. A carrier was defined as any asymptomatic person from the facility from whom erythromycin-resistant GAS was isolated from a skin or throat swab sample.
Comparison of Outbreak Isolates with Nonoutbreak Isolates
After the initial 3 cases were confirmed, throat swab samples were collected from all residents and from facility staff members who worked in nursing, kitchen, or waste collection. Swab samples were also collected from any skin lesion on staff members or residents. Hospital staff working in the wards where residents had been admitted were also asked to consent to collection of throat swab samples. The outbreak isolates were compared with nonoutbreak isolates for contextual purposes. Nonoutbreak isolates were defined as clinical isolates submitted to the Institute of Environmental Science and Research (ESR) in New Zealand from across New Zealand during 2002–2014. Although including only contemporaneous nonoutbreak isolates would have been ideal (that is, only those collected in 2014), New Zealand’s small population and the fact that GAS infections are not notifiable in New Zealand meant that we had to select nonoutbreak isolates over a broader time frame. All outbreak and nonoutbreak isolates underwent initial emm typing at ESR according to previously described methods (7). We included in our compariative analyis only nonoutbreak isolates that had the same emm type as the outbreak isolates. Data collection was approved by the Medicine, Dentistry and Health Sciences Human Ethics Sub-Committee at the University of Melbourne (ID no. 1853078).
Genome Sequencing and Assembly
We performed genome sequencing and assembly for outbreak and nonoutbreak isolates. All isolates underwent Illumina whole-genome sequencing (https://www.illumina.com). To enable fine-mapping of the outbreak, we completely sequenced a representative emm81 outbreak isolate, DMG1800716, by using Pacific Biosciences long-read technology (https://www.pacb.com). To validate the consensus assembly of the reference genome, we used Illumina short reads. We used Prokka (10) with manual curation to annotate the final sequence and SPAdes version 3.9.0 (11) for de novo assembly of raw Illumina reads into draft assemblies. We performed pairwise BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) comparisons of the 55 emm81 genomes relative to DMG1800716 by using the BLAST Ring Image Generator (12). We submitted the complete genome sequence of DMG1800716 to GenBank under accession no. CP027771. Short reads of all sequenced isolates are available at the National Center for Biotechnology Information sequence read archive (https://www.ncbi.nlm.nih.gov/sra) under BioProject PRJNA494270.
Phylogenetic Analyses
To determine whether outbreak isolates were genetically related, we mapped the genomes of the outbreak isolates and nonoutbreak isolates from across New Zealand to the newly generated 1,869,673-bp emm81 outbreak reference genome, DMG1800716 (Appendix 1). We inferred phylogenetic relationships by both maximum-likelihood and Bayesian assessment of core-genome single-nucleotide polymorphism (SNPs). We used consensus SNP alignments to build a maximum-likelihood tree with RAxML version 8.0.1 (13) and assessed temporal phylogenetic analysis by using BEAST version 2.4.7 (14), and a Hasegawa-Kishino-Yano plus gamma site model with a strict clock model after assessing temporal signal by using TempEst (15).
Clinical Epidemiology of the Outbreak
During the 2 phases of the outbreak, 14 cases of erythromycin-resistant GAS infection were confirmed: 10 in residents and 4 in staff members. Eleven of the confirmed cases (10 in residents, 1 in a staff member) were detected by swabbing of unwell persons with suspected cases, and the other 3 confirmed cases were identified by prospective sampling of all 75 residents and 30 hospital staff members (for each of these staff members, the outbreak strain was isolated from skin lesions on their hands). Prospective swabbing also identified 1 resident as a carrier (erythromycin-resistant GAS was isolated from the resident’s throat).
The average case-patient age was 79.5 years. Residents exhibited a variety of signs and symptoms (e.g., fever, malaise, suspected septic arthritis, diarrhea and vomiting, abdominal pain, and skin lesions). One staff member was hospitalized with severe pharyngitis; the other staff members were treated at home for minor skin infections.
During the first phase of the outbreak, a characteristic feature was the rapidity with which case-patient conditions deteriorated; 1 died within a few hours of symptom onset. Five confirmed case-patients, all residents, died of streptococcal sepsis during the first phase.
During the second phase of the outbreak, no deaths were reported. Five persons had suspected cases (4 residents, 1 staff member) during the outbreak but were excluded from this analysis because erythromycin-resistant GAS was not isolated.
Outbreak Isolates
During our investigation, we obtained 18 erythromycin-resistant GAS isolates, which were cultured from a variety of body sites including blood, throat, and soft tissue (Table). For 2 residents, identical outbreak strains were isolated from 2 different body sites; the remaining isolates each came from different patients. The phenotypic antimicrobial sensitivity pattern of the outbreak isolates included susceptibility to penicillin, methicillin (oxacillin), amoxicillin, and vancomycin and resistance to erythromycin with inducible resistance to clindamycin. No GAS with this antibiogram was cultured from the 65 screening throat swab samples from external hospital staff (those working in wards where case-patients were admitted).
Outbreak Management Interventions
After the initial 3 cases were confirmed and the outbreak was recognized, public health staff members initiated targeted chemoprophylaxis. A 10-day course of penicillin or amoxicillin was given to all staff members, any resident who was unwell or had been in contact with a case-patient, and any resident from whom GAS was isolated. The 4 staff members with outbreak strain infections stayed away from work until they had completed their course of antimicrobial therapy, their clinical signs of infections had resolved, and a throat swab sample culture was negative. Outbreak control measures initially continued for 1 month after the last case in the first outbreak phase was identified.
When the outbreak recurred, additional surveillance and environmental control measures were initiated and continued for 3 months after the last case of the second phase was identified. Other additional control measures included educating staff and residents about hand hygiene, monitoring the temperature of any resident with a skin lesion, cleaning all furniture and upholstery with diluted bleach where possible, replacing all toothbrushes, using disposable wound dressing trays rather than trolleys, inspecting the hands of staff members for skin lesions daily, and instructing the hospital to collect blood and throat swab samples for culture from any residents admitted from this eldercare facility and to place them in a single room. Items used communally by residents and staff (e.g., salt and pepper shakers, portable telephones) were cleaned with diluted bleach after meals or each use.
Genomic Epidemiology
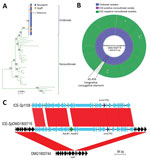
Molecular analysis of the 18 outbreak GAS strains indicated that they all contained the emm81.0 gene allele. Only 5 contemporaneous nonoutbreak emm81 isolates were collected in New Zealand during 2014; the remaining 32 nonoutbreak isolates were collected during 2002–2013. Core-genome comparisons of the 18 outbreak strains with the 37 nonoutbreak emm81 isolates showed that the outbreak isolates were highly clonal and formed a separate clade in the emm81 phylogeny (Figure, panel A). Although 336 core-genome SNPs were identified among all emm81 isolates studied, no SNPs were identified between 15 of the outbreak isolates and 1 SNP difference was identified in the remaining 3 isolates. One isolate from a staff member differed from the outbreak isolates by a single SNP in the murM locus; the remaining isolates from staff members were indistinguishable from isolates from residents (Appendix 2). These data suggest spread of the outbreak clone between staff members and residents to which directionality cannot be inferred. A single nonoutbreak isolate, DMG1800755, differed from the outbreak clade by 2 SNPs. The isolate came from an aspirate from a patient in the same southern region of New Zealand in mid-2014, around the temporal midpoint of the outbreak. The remaining nonoutbreak isolates showed a distant evolutionary relationship to the outbreak emm81 lineage.
To understand molecular differences between the outbreak emm81 lineage and the unrelated nonoutbreak emm81 isolates, we investigated genome-wide heterogeneity of the 55 emm81 genomes. We screened core genomes for mutations within the key GAS regulatory genes covR/S, ropB, mga, which had previously been linked to increased virulence among GAS isolates (1); we found no differential mutations between outbreak and nonoutbreak isolates. Comparison of the accessory (variable) genome content of the 55 emm81 isolates revealed that all isolates sampled from South Island during the outbreak period harbored an integrative conjugative element (ICE); all other nonoutbreak isolates sampled during the outbreak period, which were all from other geographic regions, were ICE negative (Figure, panel B). The outbreak ICE element, called ICE-SpDMG1800716, shared 99% nucleotide sequence homology with ICE-Sp1108, previously described for an erythromycin-resistant GAS isolate from Italy (17) (Figure, panel C). Comparative analysis revealed that ICE-SpDMG1800716 contained the inducible macrolide-resistance gene, erm(TR). ICE-SpDMG1800716 also harbored the abortive infection operon, AbiE operon, which is associated with bacteriophage resistance and stabilization of extrachromosomal elements (17). The ICE was integrated between the 3′ end of the 23s tRNA methyltransferase (rum) gene (17) and the 5′ end of a phosphorylase gene of a representative nonoutbreak isolate (DMG1800744) (Figure, panel C). Bayesian temporal analysis of the emm81 population indicated that ICE-SpDMG1800716 was acquired during 2007–2013 and that the ICE-positive clade subsequently expanded in 2013 (95% confidence range 2013–2014; Appendix 1 Figure).
Through our clinical and genomic epidemiologic analyses, we determined that a fatal GAS outbreak in an eldercare facility was associated with a single emm81 GAS clone that was resistant to erythromycin and exhibited inducible clindamycin resistance. emm81 GAS is one of the most common M-types that causes invasive disease in New Zealand (3). The role of emm81 as a global GAS strain is highlighted by its inclusion in the experimental 30-valent M-protein vaccine (19). Traditional typing methods, such as emm typing, would not have had the discriminatory power to differentiate the outbreak isolates from other emm81 isolates that were already in New Zealand. Along with other recent reports of GAS outbreaks of a single emm type (8,9), our study highlights the utility of whole-genome sequencing as an epidemiologic tool for GAS outbreak investigations.
Comparative analyses of the outbreak clone with 37 nonoutbreak emm81 isolates identified that the outbreak clone had acquired a macrolide resistance determinant within a putative integrative and conjugative element, ICE-SpDMG1800716. Outbreaks of GAS disease have previously been linked to the acquisition of mobile genetic elements, such as an ongoing polyclonal emm12 and emm1 scarlet fever outbreak in Hong Kong and mainland China associated with horizontal acquisition of multidrug resistance and a superantigen-encoding prophage (6). Outbreaks of invasive GAS disease have also been associated with acquisition of, or mutations within, genotypic regulatory systems that result in increased phenotypic virulence (20,21). However, this clonal invasive GAS outbreak differs from previously reported outbreaks (6,20,21) because it was linked primarily to the acquisition of a transposable element with no obvious virulence determinant. Widespread use of macrolides in New Zealand, particularly in elderly patients and during the winter, when this outbreak occurred (22), may well have contributed to the selection and expansion of the macrolide-resistant outbreak clone.
In addition to harboring macrolide-resistance genes, the integrative conjugative element in the outbreak isolates also contained the abortive infection protein AbiE, which may have contributed to the relative fitness of the outbreak isolates. AbiE may confer bacteriophage resistance and has been shown to stabilize extrachromosomal elements such as plasmids (23); thus, its presence may have helped maintain ICE-SpDMG1800716 within the genomes of the outbreak isolates.
GAS carriage among healthcare workers in this and other outbreaks serves as a reminder that staff member sampling is integral to GAS outbreak investigations (9). Such practices, although common in hospital settings, are not universally followed during investigations of outbreaks in long-term care or eldercare facilities (5). In addition to a geographic and temporal link between the outbreak isolates and their closest nonoutbreak relative (both being from southern New Zealand and isolated in 2014), no contact history could be determined between the clinical nonoutbreak isolate and the outbreak facility. We therefore hypothesize that the outbreak probably commenced from an unsampled community source that gained entry to the facility by contact with either a resident or staff member. Although an environmental source is unlikely, environmental sampling was not undertaken; thus, fomites such as communal dinnerware or telephones could have been the common source of infection that resulted in the second phase of the outbreak. In a review of 17 reports of GAS outbreaks in long-term care facilities, fomites were not definitively implicated in outbreak transmission (5); therefore, an environmental source indeed seems less likely as a source of the recurrence of this outbreak and an unsampled human source seems more likely.
As was the case for other reported outbreaks in long-term care facilities (8,9), improved infection control measures and chemoprophylaxis were the cornerstones of outbreak control in this outbreak. Although infection control measures are undoubtedly of utmost importance, the evidence as to whether targeted or mass chemoprophylaxis is preferable in eldercare settings is conflicting, because risk for secondary invasive GAS infection is higher among elderly persons than among other contacts (24,25). Authors of a recent UK study demonstrated a considerably increased risk for invasive GAS infection among household contacts, particularly for persons >75 years of age, for whom the fatality rate for secondary cases was 19% (24). They suggested that, even in nonoutbreak settings, targeted chemoprophylaxis for elderly household contacts of invasive GAS patients should be considered (24). It is conceivable that the targeted chemoprophylaxis undertaken during the first phase of this outbreak prevented some cases, yet the occurrence of the second phase suggests that this approach alone was not sufficient. A 2007 review of 17 GAS outbreaks in long-term care facilities similarly found that in 3 facilities, targeted chemoprophylaxis was insufficient for achieving outbreak control and that control was achieved only after the facilities initiated mass chemoprophylaxis to augment existing infection control measures (5). More recently, mass chemoprophylaxis was insufficient for halting a multiphase outbreak in 2 long-term care facilities in the United States (9). In that study, mass chemoprophylaxis was initiated for all residents and consenting staff; prophylactic coverage was wider than that in our study. Nevertheless, mass chemoprophylaxis in the US outbreak was still only partially effective, and outbreak persistence was attributed mostly to continued lapses in infection control practices. During a GAS outbreak in another long-term care facility, breaches in infection control practices were also noted; prospective assessment of staff members’ wound care and hand hygiene practices found several lapses in each (26). In our study, improved infection control practices were initiated, but direct observation of staff undertaking wound care and hand hygiene may have further helped to identify exactly where lapses might have been occurring.
In summary, our data further highlight the potential for invasive GAS to cause rapid and fatal outbreaks, particularly in closed communities such as eldercare facilities. Invasive GAS disease is not notifiable in New Zealand, nor is there mandatory surveillance for invasive GAS infections. The incidence of invasive GAS infections in New Zealand and elsewhere is particularly high among those >75 years of age (2,3). Our findings add to the growing body of evidence emphasizing the need for improved surveillance and response to invasive GAS infections in at-risk populations, particularly in countries such as New Zealand where active surveillance is not conducted.
Dr. Worthing is a postdoctoral research fellow at the Animal Parasitic Diseases Laboratory, Beltsville, MD, USA. Her research interests include using genomics to investigate antimicrobial-resistant pathogens of humans and animals in a One Health setting.
Acknowledgments
We thank the laboratory staff at the Microbiological Diagnostic Unit at the Doherty Institute for Infection and Immunity, The University of Melbourne, and the core sequencing facility at the Wellcome Trust Sanger Institute, UK. We also acknowledge Seamus O’Reilly for his assistance in reviewing this manuscript.
This work was supported by The Wellcome Trust, UK, and the National Health and Medical Research Council of Australia (grant no. 1130455), and an Early Career Fellowship (GNT1123854 to D.A.W.)
References
- Walker MJ, Barnett TC, McArthur JD, Cole JN, Gillen CM, Henningham A, et al. Disease manifestations and pathogenic mechanisms of Group A Streptococcus. Clin Microbiol Rev. 2014;27:264–301. DOIPubMedGoogle Scholar
- Nelson GE, Pondo T, Toews K-A, Farley MM, Lindegren ML, Lynfield R, et al. Epidemiology of invasive group A streptococcal infections in the United States, 2005–2012. Clin Infect Dis. 2016;63:478–86. DOIPubMedGoogle Scholar
- Williamson DA, Morgan J, Hope V, Fraser JD, Moreland NJ, Proft T, et al. Increasing incidence of invasive group A streptococcus disease in New Zealand, 2002-2012: a national population-based study. J Infect. 2015;70:127–34. DOIPubMedGoogle Scholar
- Thigpen MC, Richards CL Jr, Lynfield R, Barrett NL, Harrison LH, Arnold KE, et al.; Active Bacterial Core surveillance / Emerging Infections Program Network. Invasive group A streptococcal infection in older adults in long-term care facilities and the community, United States, 1998-2003. Emerg Infect Dis. 2007;13:1852–9. DOIPubMedGoogle Scholar
- Jordan HT, Richards CL Jr, Burton DC, Thigpen MC, Van Beneden CA. Group a streptococcal disease in long-term care facilities: descriptive epidemiology and potential control measures. Clin Infect Dis. 2007;45:742–52. DOIPubMedGoogle Scholar
- Davies MR, Holden MT, Coupland P, Chen JH, Venturini C, Barnett TC, et al. Emergence of scarlet fever Streptococcus pyogenes emm12 clones in Hong Kong is associated with toxin acquisition and multidrug resistance. Nat Genet. 2015;47:84–7. DOIPubMedGoogle Scholar
- Beall B, Facklam R, Thompson T. Sequencing emm-specific PCR products for routine and accurate typing of group A streptococci. J Clin Microbiol. 1996;34:953–8. DOIPubMedGoogle Scholar
- Chalker VJ, Smith A, Al-Shahib A, Botchway S, Macdonald E, Daniel R, et al. Integration of genomic and other epidemiologic data to investigate and control a cross-institutional outbreak of Streptococcus pyogenes. Emerg Infect Dis. 2016;22:973–80. DOIPubMedGoogle Scholar
- Nanduri SA, Metcalf BJ, Arwady MA, Edens C, Lavin MA, Morgan J, et al. Prolonged and large outbreak of invasive group A Streptococcus disease within a nursing home: repeated intrafacility transmission of a single strain. Clin Microbiol Infect. 2019;25:248.e1–7. DOIPubMedGoogle Scholar
- Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–9. DOIPubMedGoogle Scholar
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–77. DOIPubMedGoogle Scholar
- Alikhan N-F, Petty NK, Ben Zakour NL, Beatson SA. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. 2011;12:402. DOIPubMedGoogle Scholar
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–3. DOIPubMedGoogle Scholar
- Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu C-H, Xie D, et al. BEAST 2: a software platform for Bayesian evolutionary analysis. PLOS Comput Biol. 2014;10:
e1003537 . DOIPubMedGoogle Scholar - Rambaut A, Lam TT, Max Carvalho L, Pybus OG. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen). Virus Evol. 2016;2:
vew007 . DOIPubMedGoogle Scholar - Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44(W1):
W242-5 . DOIPubMedGoogle Scholar - Brenciani A, Tiberi E, Bacciaglia A, Petrelli D, Varaldo PE, Giovanetti E. Two distinct genetic elements are responsible for erm(TR)-mediated erythromycin resistance in tetracycline-susceptible and tetracycline-resistant strains of Streptococcus pyogenes. Antimicrob Agents Chemother. 2011;55:2106–12. DOIPubMedGoogle Scholar
- Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer. Bioinformatics. 2011;27:1009–10. DOIPubMedGoogle Scholar
- Dale JB, Penfound TA, Chiang EY, Walton WJ. New 30-valent M protein-based vaccine evokes cross-opsonic antibodies against non-vaccine serotypes of group A streptococci. Vaccine. 2011;29:8175–8. DOIPubMedGoogle Scholar
- Cornick JE, Kiran AM, Vivancos R, Van Aartsen J, Clarke J, Bevan E, et al. Epidemiological and molecular characterization of an invasive group A Streptococcus emm32.2 outbreak. J Clin Microbiol. 2017;55:1837–46. DOIPubMedGoogle Scholar
- Galloway-Peña J, Clement ME, Sharma Kuinkel BK, Ruffin F, Flores AR, Levinson H, et al. Application of whole-genome sequencing to an unusual outbreak of invasive group A streptococcal disease. [-ofw.]. Open Forum Infect Dis. 2016;3:
ofw042 . DOIPubMedGoogle Scholar - Williamson DA, Roos R, Verrall A, Smith A, Thomas MG. Trends, demographics and disparities in outpatient antibiotic consumption in New Zealand: a national study. J Antimicrob Chemother. 2016;71:3593–8. DOIPubMedGoogle Scholar
- Dy RL, Przybilski R, Semeijn K, Salmond GPC, Fineran PC. A widespread bacteriophage abortive infection system functions through a Type IV toxin-antitoxin mechanism. Nucleic Acids Res. 2014;42:4590–605. DOIPubMedGoogle Scholar
- Mearkle R, Saavedra-Campos M, Lamagni T, Usdin M, Coelho J, Chalker V, et al. Household transmission of invasive group A Streptococcus infections in England: a population-based study, 2009, 2011 to 2013. Euro Surveill. 2017;22:30532. DOIPubMedGoogle Scholar
- Adebanjo T, Apostol M, Alden N, Petit S, Tunali A, Torres S, et al. Evaluating household transmission of invasive group A Streptococcus disease in the United States using population-based surveillance data, 2013–2016. Clin Infect Dis. 2019 Aug 13;(ciz716). Epub ahead of print.
- Ahmed SS, Diebold KE, Brandvold JM, Ewaidah SS, Black S, Ogundimu A, et al. The role of wound care in 2 group A streptococcal outbreaks in a Chicago skilled nursing facility, 2015‒2016. Open Forum Infect Dis. 2018;5:
ofy145 . DOIPubMedGoogle Scholar
Figure
Table
Cite This ArticleOriginal Publication Date: April 01, 2020
1These authors contributed equally to this article.
Table of Contents – Volume 26, Number 5—May 2020
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Mark R. Davies, Department of Microbiology and Immunology, The University of Melbourne at the Peter Doherty Institute for Infection and Immunity, 792 Elizabeth St, Melbourne, Victoria 3000, Australia
Top