Volume 26, Number 7—July 2020
Dispatch
Carbapenem Resistance Conferred by OXA-48 in K2-ST86 Hypervirulent Klebsiella pneumoniae, France
Cite This Article
Citation for Media
Abstract
We recovered 2 carbapenem-resistant K2-ST86 hypermucoviscous Klebsiella pneumoniae isolates from patients in France. The isolates had genetic attributes of hypervirulent K. pneumoniae but differed in ability to cause mouse lethality. Convergence of hypervirulent K. pneumoniae toward resistance could cause a health crisis because such strains could be responsible for severe and untreatable infections.
Klebsiella pneumoniae is a threat to human health because of the emergence of hypervirulent K. pneumoniae, which has caused severe community-acquired infections, and classical multidrug-resistant K. pneumoniae involved in hospital outbreaks (1). Classical K. pneumoniae generally lacks the virulence genes associated with invasive diseases (1) and belongs to successful clonal groups, such as sequence type (ST) 11 and ST258 (2). Most hypervirulent K. pneumoniae isolates, which are mainly found in Asia (3,4), belong to the K1 and K2 capsular serotypes and are restricted to clonal complexes different from classical multidrug-resistant K. pneumoniae groups, such as K1-ST23, the most prevalent group (2). They rarely harbor acquired antimicrobial resistance genes but have virulence loci and a hypermucoviscous phenotype (5). We describe 2 hypermucoviscous K2-ST86 K. pneumoniae (positive string test) resistant to carbapenems isolated in northern and southern France.
In 2017, we recovered the Kpn154 strain from the urine of a 35-year-old man with community-acquired urinary tract infection. He had fever (39°C) before local symptoms suggesting urinary tract infection caused by bacteremic spread, which was successfully treated with intravenous ceftriaxone. A second strain, Kpn2166, was hospital-acquired and recovered from the feces of a 70-year-old man in the intensive care unit of the hospital at which the 35-year-old patient was seem. Neither patient reported travel during the past 4 years. Both strains were resistant to all penicillins and their combinations with β-lactamase inhibitors, and to carbapenems according to EUCAST (European Committee on Antimicrobial Susceptibility Testing) guidelines (https://www.eucast.org) (Table). In addition, Kpn2166 was resistant to the third-generation cephalosporins, quinolones and tigecycline.
We obtained the isolates’ whole-genome sequence by hybrid de novo assembly of short and long reads generated with technologies from Illumina (https://www.illumina.com) and Oxford Nanopore (https://nanoporetech.com; European Nucleotide Archive at EMBL-EBI under accession no. PRJEB34867). We typed the isolates as K2-ST86 from whole-genome sequencing using the Institut Pasteur multilocus sequence typing scheme (https://bigsdb.pasteur.fr) and Kleborate (6). Kpn154 harbored carbapenemase-encoding gene blaoxa-48 and Kpn2166 the extended-spectrum β-lactamase–encoding gene blaCTX-M-15 as the only acquired β-lactamase–encoding genes. CTX-M-15 associated with the truncation of the outer membrane protein OmpK36 caused by 11-bp deletion was probably responsible for carbapenem resistance in Kpn2166. In the absence of rpsJ variants previously associated with tigecycline resistance, Kpn2166 resistance to tigecycline and quinolones probably resulted from a frameshift mutation in ramR (C→T at position 364) (7).
We identified IncL replicon in Kpn154 and IncN replicon in Kpn2166. The IncL replicon of Kpn154 was a canonical pOXA-48–like plasmid encoding blaOXA-48 (GenBank accession no. JN626286). In the Kpn2166 isolate, blaCTX-M-15 was encoded by a new ST9-IncN plasmid, included in an IS26-based composite transposon and downstream a truncated ISEcp1 insertion sequence.
Each isolate harbored an IncHI1B/IncFIB replicon, designated pVIR-Kpn154 and pVIR-Kpn2166 (Table), typified by the reference hvKP virulence plasmid pLVPK (8). They shared with pLVPK 97% pairwise identity overall and all virulence genes, including the rmpA and rmpA2 genes involved in the hypermucoid phenotype (8). We typed the rmpA genes as allele 2 according to the Institut Pasteur scheme. However, Kpn2166 rmpA2 harbored in addition a frameshift mutation (coding sequence [CDS] position 196) and 2 other mutations in virulence genes encoding the siderophores (Appendix Figure 1).
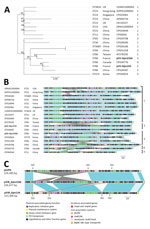
We compared the pVIR-Kpn154 and pVIR-Kpn2166 plasmids with 16 complete hypervirulent K. pneumoniae plasmid sequences (Appendix Table) from the PATRIC database (http://www.patricbrc.org). A single-nucleotide polymorphism–based phylogenic tree showed a link between the major tree branches and the sequence types of hypervirulent K. pneumoniae owners but not with their geographic origin (Figure 1, panel A). This finding suggests emergence was more likely caused by spread over distant geographic areas than by local expansion and that limited horizontal transfers between hypervirulent K. pneumoniae isolates probably resulted from the absence of known genes involved in conjugation. Phylogenetic tree analysis also showed 5 groups based on virulence gene synteny, with the predominant group typified by pLVPK (Figure 1, panel B). Because plasmids of ST23-like hypervirulent K. pneumoniae all share a similar synteny of virulence genes, ST86 hypervirulent K. pneumoniae contains a diversity of plasmid synteny groups (Figure 1, panel A), suggesting that rearrangements of virulence genes occurred several times along plasmid evolution at rearrangement hotspots active in non-ST23 genetic background. For example, pVIR-Kpn2166 and pVIR-Kpn154 differed from reference plasmid pLVPK by the permutations in the ≈100-kb region flanked by IS5 mobile elements (Figure 1, panel C). pVIR-Kpn154 contained an additional copy of IS5, which was associated with another ≈30-kb permutation + translation event, suggesting that IS5 is a key factor in the evolution and diversity of hypervirulent K. pneumoniae plasmids.
The chromosome of Kpn154 and Kpn2166 exhibited similar organization but differed by 128 insertion/deletion mutations and 1,928 single-nucleotide variants (Appendix Figures 2, 3). The Kpn154 chromosome-mediated ybt virulence locus, which encodes the yersiniabactin, was located in the integrative conjugative element ICEKp3 and was typed ybST202–1LV (6). In Kpn2166, ybt was located in an original isoform of ICEKp12, presenting an ≈34-kb deletion compared to the canonical 97,771-bp ICEKp12, and was typed ybST13–1LV (6).
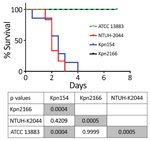
Although Kpn154 and Kpn2166 have the same genetic background and share the same virulence score (6), they also have allelic and synteny differences in virulence genes. We therefore compared the virulence of these isolates in a sepsis model based on outbred mice challenged intraperitoneally, as described (Figure 2) (10). Mice injected with 103 CFUs of Kpn154 or hypervirulent K. pneumoniae reference strain K. pneumoniae NTUH-2044 died in <72 h, in contrast to mice inoculated with Kpn2166 or American Type Culture Collection (ATCC) 13883, showing that only isolate Kpn154 is hypervirulent. Higher bacterial doses (106 and 108 CFUs) of ATCC13883 and Kpn2166 did not lead to mouse lethality, confirming that Kpn2166 is not hypervirulent in this model, despite harboring all genetic attributes of hypervirulent K. pneumoniae except a functional rmpA2 gene and allelic variants of siderophore-encoding genes.
We assessed the production of siderophores in Kpn154, Kpn2166 and the control strains as described (11). Although the siderophore production of the nonvirulent strain ATCC13883 (mean 30.2 + SD 1.8 μg/mL) was at the previously reported rate predicting hypervirulent K. pneumoniae phenotype (≥30 μg/mL), the other strains produced significantly higher siderophore levels (107.2 + 4.5 μg/mL to 306.7 + 20.2 μg/mL; Bonferroni-adjusted p = 0.0035 by Mann-Whitney test), with Kpn154 producing at the lower level (Appendix Figure 4).
Our results show that a hypermucoviscous K2-ST86 strain can be avirulent in a sepsis mouse model and that hypervirulence cannot be clearly explained by siderophore production alone. Gene rmpA2, not required for the hypermucoviscosity phenotype as previously observed (12), might be required for hypervirulent phenotype because it is a main, but not the only, difference we observed between the ST86-K2 strains. Finally, these results highlight the importance of in vivo virulence investigation to identify hypervirulent K. pneumoniae, especially in the absence of an appropriate clinical scenario.
The threat of hypervirulent K. pneumoniae acquiring carbapenem resistance is becoming a reality in Asia, especially in China, where hypervirulence prevalence among carbapenem-resistant K. pneumoniae is 7.4%–15% (5). Most resistant isolates are non-K1/K2-ST11 and produce carbapenemase KPC-2;. they result from the transfer of the pLVPK-like plasmid into ST11 classical multidrug-resistant K. pneumoniae isolates, as observed in the 2 cases reported outside China (5). Inversely, the carbapenemase-producing isolate Kpn154 results from the transformation of K2-ST86 hypervirulent K. pneumoniae by plasmid encoding carbapenemase OXA-48, the most prevalent carbapenemase in France. Similar events occurred with the KPC-2 K2-ST86 isolate recently reported in Canada (13) and a few KPC-2 and NDM K1-ST23 cases documented in China and recently in the United States and United Kingdom (5,14,15). The combination of multidrug resistance and enhanced virulence has the potential to trigger the next clinical crisis and cause severe and untreatable infections in previously healthy persons.
Dr. Beyrouthy is a microbiologist in the Associated French National Reference Center for Antibiotic Resistance, Clermont-Ferrand, Clermont-Ferrand University Hospital, and a member of the Microbe, Intestine, Inflammation and Host Susceptibility (M2ISH) research group of the Institut National de la Santé et de la Recherche Médicale. Her primary research interests are antimicrobial drug resistance and Enterobacteriaceae epidemiology.
Acknowledgments
We thank Alexis Pontvianne and Lucie Pourpuech for their technical assistance.
This work was supported by the Ministere de l’Education Nationale, de l’Enseignement Superieur et de la Recherche, Institut National de la Santé et de la Recherche Médicale (UMR1071), INRA (USC-2018), Santé Publique France, and the Centre Hospitalier Regional Universitaire de Clermont-Ferrand, France.
References
- Meatherall BL, Gregson D, Ross T, Pitout JDD, Laupland KB. Incidence, risk factors, and outcomes of Klebsiella pneumoniae bacteremia. Am J Med. 2009;122:866–73. DOIPubMedGoogle Scholar
- Wyres KL, Wick RR, Judd LM, Froumine R, Tokolyi A, Gorrie CL, et al. Distinct evolutionary dynamics of horizontal gene transfer in drug resistant and virulent clones of Klebsiella pneumoniae. PLoS Genet. 2019;15:
e1008114 . DOIPubMedGoogle Scholar - Prokesch BC, TeKippe M, Kim J, Raj P, TeKippe EME, Greenberg DE. Primary osteomyelitis caused by hypervirulent Klebsiella pneumoniae. Lancet Infect Dis. 2016;16:e190–5. DOIPubMedGoogle Scholar
- Shon AS, Bajwa RPS, Russo TA. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence. 2013;4:107–18. DOIPubMedGoogle Scholar
- Lee CR, Lee JH, Park KS, Jeon JH, Kim YB, Cha CJ, et al. Antimicrobial resistance of hypervirulent Klebsiella pneumoniae: epidemiology, hypervirulence-associated determinants, and resistance mechanisms. Front Cell Infect Microbiol. 2017;7:483. DOIPubMedGoogle Scholar
- Lam MMC, Wick RR, Wyres KL, Gorrie CL, Judd LM, Jenney AWJ, et al. Genetic diversity, mobilisation and spread of the yersiniabactin-encoding mobile element ICEKp in Klebsiella pneumoniae populations. Microb Genom. 2018;4. DOIPubMedGoogle Scholar
- Villa L, Feudi C, Fortini D, García-Fernández A, Carattoli A. Genomics of KPC-producing Klebsiella pneumoniae sequence type 512 clone highlights the role of RamR and ribosomal S10 protein mutations in conferring tigecycline resistance. Antimicrob Agents Chemother. 2014;58:1707–12. DOIPubMedGoogle Scholar
- Chen YT, Chang HY, Lai YC, Pan CC, Tsai SF, Peng HL. Sequencing and analysis of the large virulence plasmid pLVPK of Klebsiella pneumoniae CG43. Gene. 2004;337:189–98. DOIPubMedGoogle Scholar
- Beyrouthy R, Barets M, Marion E, Dananché C, Dauwalder O, Robin F, et al. Novel enterobacter lineage as leading cause of nosocomial outbreak involving carbapenemase-producing strains. Emerg Infect Dis. 2018;24:1505–15. DOIPubMedGoogle Scholar
- Beyrouthy R, Robin F, Cougnoux A, Dalmasso G, Darfeuille-Michaud A, Mallat H, et al. Chromosome-mediated OXA-48 carbapenemase in highly virulent Escherichia coli. J Antimicrob Chemother. 2013;68:1558–61. DOIPubMedGoogle Scholar
- Russo TA, Olson R, Fang CT, Stoesser N, Miller M, MacDonald U, et al. Identification of biomarkers for differentiation of hypervirulent Klebsiella pneumoniae from classical K. pneumoniae. J Clin Microbiol. 2018;56:1–12. DOIPubMedGoogle Scholar
- Hsu CR, Lin TL, Chen YC, Chou HC, Wang JT. The role of Klebsiella pneumoniae rmpA in capsular polysaccharide synthesis and virulence revisited. Microbiology. 2011;157:3446–57. DOIPubMedGoogle Scholar
- Mataseje LF, Boyd DA, Mulvey MR, Longtin Y. Two hypervirulent Klebsiella pneumoniae isolates producing a blaKPC-2 carbapenemase from a Canadian patient. Antimicrob Agents Chemother. 2019;63:1–4. DOIPubMedGoogle Scholar
- Karlsson M, Stanton RA, Ansari U, McAllister G, Chan MY, Sula E, et al. Identification of a carbapenemase-producing hypervirulent Klebsiella pneumoniae isolate in the United States. Antimicrob Agents Chemother. 2019;63:1–6. DOIPubMedGoogle Scholar
- Roulston KJ, Bharucha T, Turton JF, Hopkins KL, Mack DJF. A case of NDM-carbapenemase-producing hypervirulent Klebsiella pneumoniae sequence type 23 from the UK. JMM Case Rep. 2018;5:
e005130 . DOIPubMedGoogle Scholar
Figures
Table
Cite This ArticleOriginal Publication Date: June 15, 2020
Table of Contents – Volume 26, Number 7—July 2020
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Racha Beyrouthy, Centre Hospitalier Universitaire, Centre de Biologie, Service de Bactériologie, 58 rue Montalembert, 63000 Clermont-Ferrand, France
Top