Volume 26, Number 9—September 2020
Dispatch
Sequence Type Changes Associated with Decreasing Macrolide-Resistant Mycoplasma pneumoniae, Japan
Cite This Article
Citation for Media
Abstract
We compared sequence types (STs) of Mycoplasma pneumoniae isolates from Japan during 2002–2019. ST3 and ST14 dominated during 2002–2016, and ST7 and ST33 dominated during 2018–2019. These STs were associated with a decrease in macrolide-resistant strains after an epidemic of infection with M. pneumoniae during 2011–2012.
Mycoplasma pneumoniae is a major cause of community-acquired pneumonia, and macrolide-resistant M. pneumoniae is a serious concern in Asia (1–3). Throughout Japan, an outbreak of macrolide-resistant M. pneumoniae infection occurred during 2011–2012 (2). After this outbreak, the number of drug-resistant strains decreased for every year from 2013 through 2019. In contrast, China and South Korea still showed a high rate of macrolide resistance in M. pneumoniae during 2014–2018 (1,3). We determined antimicrobial drug susceptibility and performed analysis by using multilocus sequence typing (MLST), clonal complexes (CCs), and P1 gene typing for M. pneumoniae isolated from children to identify trends concerning this bacterium in Japan.
We obtained nasopharyngeal swab samples from patients who had pneumonia or bronchitis at 21 medical institutions throughout Japan during October 2018–July 2019. We collected samples after obtaining informed consent from patients or their family members (Ethics Committee approval no. 2016–0015, Keio University School of Medicine, Tokyo, Japan).
We suspended samples in 0.5 mL of pleuropneumonia-like organism broth (Difco, https://www.fishersci.com). We then performed DNA extraction by using a described protocol (4). We used a Cycleave PCR Kit (Takara Bio, https://www.takarabio.com) to detect M. pneumoniae. For confirmed cases of infection with M. pneumoniae, we used a Cycleave PCR to distinguish between macrolide-susceptible and macrolide-resistant strains (5). Cultures were grown in pleuropneumonia-like organism broth, according to previously described methods (6).
We determined MICs for antimicrobial resistance of isolates by using microdilution methods (6). We performed MLST analysis based upon sequencing of 8 housekeeping genes (ppa, pgm, gyrB, gmk, glyA, atpA, arcC, and adk) according to the method described in the MLST database (https://pubmlst.org/mpneumoniae). To determine relationships between sequence types (STs), we performed CC analysis by using global optimal eBURST (http://www.phyloviz.net/goeburst). Typing of the P1 adhesin gene in M. pneumoniae was performed as described (7).
During the 2018–2019 study period, 105 samples were received (mean patient age 8 years). M. pneumoniae was confirmed by real-time PCR in 83 (79.0%), and culturing was successful in 53 (50.5%). Of these 53 isolates, only 6 (11.3%) were macrolide-resistant M. pneumoniae. All of these macrolide-resistant strains had an A2063G mutation in the 23S rRNA gene.
We provide yearly changes in macrolide-resistant M. pneumoniae during 2002–2019 (except for 2014 and 2017) in Japan (Table). Data from the earlier years beginning in 2002 were reported previously (2,6,7). Our study group results from the earlier periods indicated macrolide-resistance rates of 6.9% (18/259) during 2002–2005; a total of 37.4% (96/257) during 2006–2009; a total of 86.2% (281/326) during 2010–2013, including the epidemic years 2011–2012; and 56.3% (111/197) during 2015–2016 compared with 11.3% (6/53) during 2018–2019. These resistance rates have decreased rapidly beginning in 2018, and the MICs for quinolone and tetracycline have remained unchanged; no drug-resistant strains were identified.
We determined relationships observed between STs and 279 macrolide-susceptible M. pneumoniae versus 191 macrolide-resistant M. pneumoniae during 2002–2019 (Figure 1). ST3 and ST14 accounted for most macrolide-susceptible M. pneumoniae during 2002–2016; these STs have been largely replaced by ST7 (n = 30, 56.6%) and ST33 (n = 13, 24.5%) during 2018–2019. ST33 first appeared in this study in 2018, and ST14 was more prevalent among macrolide-susceptible M. pneumoniae during 2002–2016 but was rarely detected during 2018–2019. Differences in STs during 2002–2016 and during 2018–2019 were highly significant (p<0.001). Conversely, most macrolide-resistant M. pneumoniae isolates belonged to ST3, as in previous years.
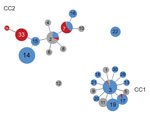
We determined relationships between CC and STs according to goeBURST (Figure 2). The data include 470 strains from our study and 62 strains registered in the MLST database from other countries. Of the 2 CC clusters, the CC1 centered on ST3, and CC2 centered on ST2. ST7 and ST33, which were prevalent during 2018–2019, belonged to CC2. Although ST14 was the most prevalent member of CC2 until 2016, ST33 replaced it and increased during 2018. Both ST14 and ST33 were derived from ST15 and showed a single-locus variant of the adk gene. We registered ST34, which was derived from ST33, as a new ST.
Results for P1 typing of M. pneumoniae showed that STs belonging to CC1 were type 1 and STs belong to CC2 were type 2. ST14, ST15, ST33, and ST34 belonged to type 2a, a subtype of P1 type 2.
In Japan, prevalence of macrolide-resistant M. pneumoniae has decreased recently and rapidly. Other study groups have reported similar trends (8). However, in countries in Asia other than Japan, the resistance rate has remained high in China (3) and South Korea (1). In the European Union, the overall rate is low, but has varied by country. Macrolide-resistant M. pneumoniae was not detected in Sweden during 1996–2013 (9), and the rate has been consistently low in Germany (1.9%–3.6%) (10). Because of tight control of antimicrobial drug prescriptions, Sweden shows extremely low use of macrolides (11) compared with for more frequent use in countries in Asia (12), where excessive use of macrolides is likely to affect selection and increase of drug-resistant strains.
In Japan, macrolide consumption has decreased gradually after the 2011–2012 outbreak of macrolide-resistant M. pneumoniae infections (http://amrcrc.ncgm.go.jp/surveillance/index.html), which may have contributed to the decrease in drug-resistant strains. In addition, the outbreak was followed by approval of tosufloxacin, a quinolone agent, for children with macrolide-resistant M. pneumoniae infection who fail to respond clinically to macrolides within 3 days. Approval of tosufloxacin as a treatment for these M. pneumoniae infections might have also contributed to the decrease in the macrolide resistance rate.
Our MLST results showed that predominant STs during 2018–2019 differed from those during 2016. M. pneumoniae could not be collected for our survey during 2017. This factor was a serious limitation because changes in STs might have occurred at that time. During 2018, strains identified as ST7 and ST33 were almost all susceptible and represented a major difference from previous findings. ST33, which contained a single-locus variant of the adk gene in ST15, was identified in Japan during 2018 and displaced ST14, the major ST among susceptible strains isolated during 2015–2016. The cause of ST replacement might have been acquisition of specific antibodies against epidemic M. pneumoniae in children and teenagers, which gave rise to a single-locus variant in its place.
CC1 is a circular clonal complex that spread from ST3 at the center toward different STs showing a single-locus variant. CC2 has extended from its original center at ST2 to include other STs with 1 or more mutated allele. STs belonging to CC2 seen to be more diverse than STs of CC1. Diversification has also been observed in type 2 of the P1 gene, corresponding to CC2 (13).
In conclusion, STs in M. pneumoniae isolates differed by area and year in Japan (1,7,14). Thus, MLST analysis is helpful in understanding worldwide trends among pathogenic M. pneumoniae.
Dr. Morozumi is an assistant professor at Keio University School of Medicine, Tokyo, Japan. Her research interests include molecular epidemiology and severe infections caused by Streptococcus agalactiae and M. pneumoniae.
Acknowledgment
We thank the pediatricians who actively participated in the M. pneumoniae Study Group (Toshiyuki Hikita, Junichi Ishikawa, Hirokazu Kutsuma, Masaaki Kobayashi, Eiichi Nakayama, Keita Matsubara, Osamu Kakuta, Kazuoki Kubota, Yosuke Mori, Hiroyoshi Ebara, Satoshi Koyama, Hiroaki Shiraishi, Shigeru Ohnari, Meiwa Shibata, Hidehisa Shinohara, Kensuke Nagai, Yasuo Kondo, and Takashige Okada); and Madoka Naito and Shinji Masuyoshi for providing laboratory assistance.
References
- Lee JK, Lee JH, Lee H, Ahn YM, Eun BW, Cho EY, et al. Clonal expansion of macrolide-resistant sequence type 3 Mycoplasma pneumoniae, South Korea. Emerg Infect Dis. 2018;24:1465–71. DOIPubMedGoogle Scholar
- Okada T, Morozumi M, Tajima T, Hasegawa M, Sakata H, Ohnari S, et al. Rapid effectiveness of minocycline or doxycycline against macrolide-resistant Mycoplasma pneumoniae infection in a 2011 outbreak among Japanese children. Clin Infect Dis. 2012;55:1642–9. DOIPubMedGoogle Scholar
- Zhao F, Li J, Liu J, Guan X, Gong J, Liu L, et al. Antimicrobial susceptibility and molecular characteristics of Mycoplasma pneumoniae isolates across different regions of China. Antimicrob Resist Infect Control. 2019;8:143. DOIPubMedGoogle Scholar
- Morozumi M, Nakayama E, Iwata S, Aoki Y, Hasegawa K, Kobayashi R, et al. Simultaneous detection of pathogens in clinical samples from patients with community-acquired pneumonia by real-time PCR with pathogen-specific molecular beacon probes. J Clin Microbiol. 2006;44:1440–6. DOIPubMedGoogle Scholar
- Liu Y, Ye X, Zhang H, Wu Z, Xu X. Rapid detection of Mycoplasma pneumoniae and its macrolide-resistance mutation by Cycleave PCR. Diagn Microbiol Infect Dis. 2014;78:333–7. DOIPubMedGoogle Scholar
- Morozumi M, Hasegawa K, Kobayashi R, Inoue N, Iwata S, Kuroki H, et al. Emergence of macrolide-resistant Mycoplasma pneumoniae with a 23S rRNA gene mutation. Antimicrob Agents Chemother. 2005;49:2302–6. DOIPubMedGoogle Scholar
- Ando M, Morozumi M, Adachi Y, Ubukata K, Iwata S. Multilocus sequence typing of Mycoplasma pneumoniae, Japan, 2002–2016. Emerg Infect Dis. 2018;24:1895–901. DOIPubMedGoogle Scholar
- Tanaka T, Oishi T, Miyata I, Wakabayashi S, Kono M, Ono S, et al. Macrolide-resistant Mycoplasma pneumoniae infection, Japan, 2008–2015. Emerg Infect Dis. 2017;23:1703–6. DOIPubMedGoogle Scholar
- Gullsby K, Bondeson K. No detection of macrolide-resistant Mycoplasma pneumoniae from Swedish patients, 1996-2013. Infect Ecol Epidemiol. 2016;6:31374. DOIPubMedGoogle Scholar
- Dumke R, Ziegler T. Long-term low rate of macrolide-resistant Mycoplasma pneumoniae strains in Germany. Antimicrob Agents Chemother. 2019;63:e00455–19. DOIPubMedGoogle Scholar
- Adriaenssens N, Coenen S, Versporten A, Muller A, Minalu G, Faes C, et al.; ESAC Project Group. European Surveillance of Antimicrobial Consumption (ESAC): outpatient macrolide, lincosamide and streptogramin (MLS) use in Europe (1997-2009). J Antimicrob Chemother. 2011;66(Suppl 6):vi37–45. DOIPubMedGoogle Scholar
- Wushouer H, Tian Y, Guan XD, Han S, Shi LW. Trends and patterns of antibiotic consumption in China’s tertiary hospitals: Based on a 5 year surveillance with sales records, 2011-2015. PLoS One. 2017;12:
e0190314 . DOIPubMedGoogle Scholar - Gullsby K, Olsen B, Bondeson K. Molecular typing of Mycoplasma pneumoniae strains in Sweden from 1996 to 2017 and the emergence of a new P1 cytadhesin gene, variant 2e. J Clin Microbiol. 2019;57:57. DOIPubMedGoogle Scholar
- Brown RJ, Nguipdop-Djomo P, Zhao H, Stanford E, Spiller OB, Chalker VJ. Mycoplasma pneumoniae epidemiology in England and Wales: a national perspective. Front Microbiol. 2016;7:157. DOIPubMedGoogle Scholar
Figures
Table
Cite This ArticleOriginal Publication Date: July 29, 2020
Table of Contents – Volume 26, Number 9—September 2020
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Kimiko Ubukata, Department of Infectious Diseases, Keio University School of Medicine, 35 Shinanomachi, Shinjuku-ku, Tokyo 160-8582, Japan
Top