Volume 27, Number 12—December 2021
Research
Mammarenaviruses of Rodents, South Africa and Zimbabwe
Figure 4
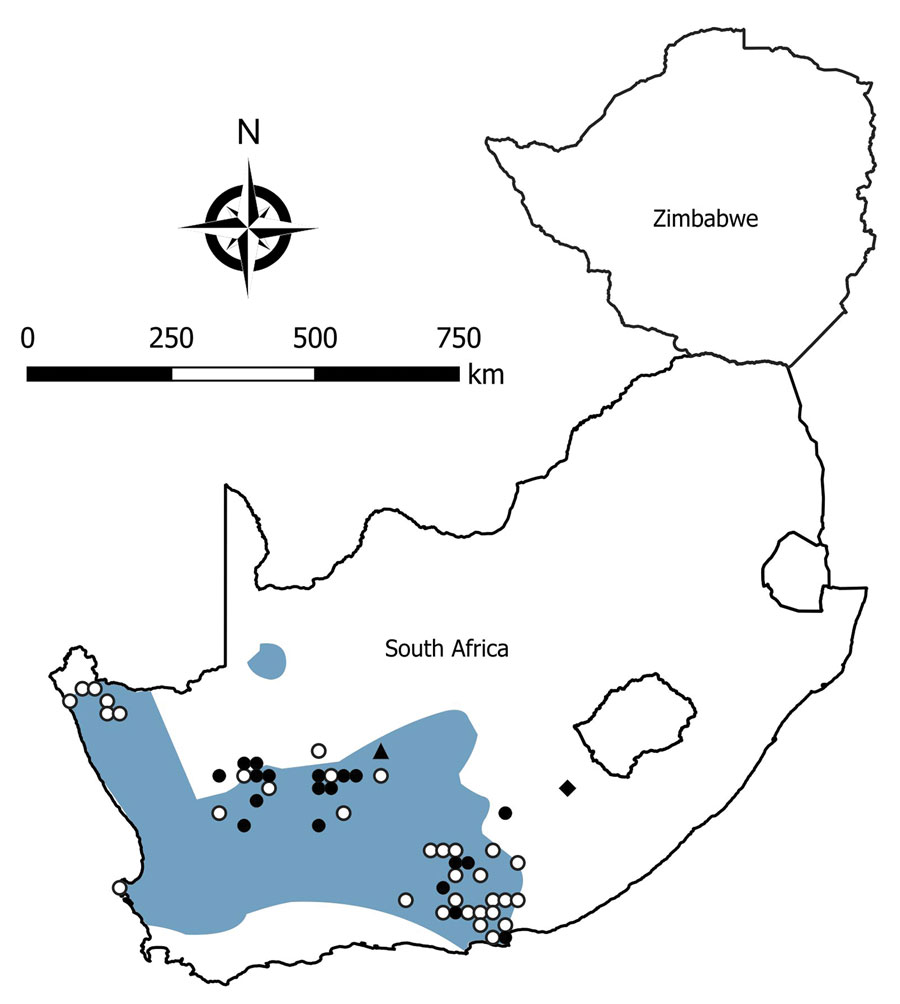
Figure 4. Locations where samples were collected from Otomys unisulcatus rodents, South Africa and Zimbabwe. White circles indicate sites where no antibody to mammarenaviruses was found in O. unisulcatus rat serum specimens; black circles, where antibody was detected in O. unisulcatus rat serum specimens; black triangle, where Omdraaivlei mammarenavirus isolates were obtained from O. unisulcatus rat samples; black diamond, where Merino Walk virus was isolated from O. unisulcatus rat. Shading indicates distribution range of O. unisulcatus rats. Adapted from Chimimba and Bennett (15).
References
- Swanepoel R. Viral haemorrhagic fevers in South Africa: history and national strategy. S Afr J Sci. 1987;83:80–8.
- Swanepoel R, Shepherd AJ, Leman PA, Shepherd SP, McGillivray GM, Erasmus MJ, et al. Epidemiologic and clinical features of Crimean-Congo hemorrhagic fever in southern Africa. Am J Trop Med Hyg. 1987;36:120–32. DOIPubMedGoogle Scholar
- Shepherd AJ, Swanepoel R, Shepherd SP, McGillivray GM, Searle LA. Antibody to Crimean-Congo hemorrhagic fever virus in wild mammals from southern Africa. Am J Trop Med Hyg. 1987;36:133–42. DOIPubMedGoogle Scholar
- Johnson KM, Elliott LH, Heymann DL. Preparation of polyvalent viral immunofluorescent intracellular antigens and use in human serosurveys. J Clin Microbiol. 1981;14:527–9. DOIPubMedGoogle Scholar
- Ksiazek TG, West CP, Rollin PE, Jahrling PB, Peters CJ. ELISA for the detection of antibodies to Ebola viruses. J Infect Dis. 1999;179(Suppl 1):S192–8. DOIPubMedGoogle Scholar
- Blackburn NK, Besselaar TG, Shepherd AJ, Swanepoel R. Preparation and use of monoclonal antibodies for identifying Crimean-Congo hemorrhagic fever virus. Am J Trop Med Hyg. 1987;37:392–7. DOIPubMedGoogle Scholar
- Swanepoel R, Blackburn NK, Efstratiou S, Condy JB. Studies on Rift Valley fever in some African murids (Rodentia: Muridae). J Hyg (Lond). 1978;80:183–96. DOIPubMedGoogle Scholar
- Galan M, Pagès M, Cosson J-F. Next-generation sequencing for rodent barcoding: species identification from fresh, degraded and environmental samples. PLoS One. 2012;7:
e48374 . DOIPubMedGoogle Scholar - Wulff H, McIntosh BM, Hamner DB, Johnson KM. Isolation of an arenavirus closely related to Lassa virus from Mastomys natalensis in south-east Africa. Bull World Health Organ. 1977;55:441–4.PubMedGoogle Scholar
- Johnson KM, Taylor P, Elliott LH, Tomori O. Recovery of a Lassa-related arenavirus in Zimbabwe. Am J Trop Med Hyg. 1981;30:1291–3. DOIPubMedGoogle Scholar
- Lecompte E, ter Meulen J, Emonet S, Daffis S, Charrel RN. Genetic identification of Kodoko virus, a novel arenavirus of the African pigmy mouse (Mus Nannomys minutoides) in West Africa. Virology. 2007;364:178–83. DOIPubMedGoogle Scholar
- de Bellocq JG, Borremans B, Katakweba A, Makundi R, Baird SJ, Becker-Ziaja B, et al. Sympatric occurrence of 3 arenaviruses, Tanzania. Emerg Infect Dis. 2010;16:692–5. DOIPubMedGoogle Scholar
- Bowen MD, Peters CJ, Nichol ST. The phylogeny of New World (Tacaribe complex) arenaviruses. Virology. 1996;219:285–90. DOIPubMedGoogle Scholar
- Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–4. DOIPubMedGoogle Scholar
- Chimimba CT, Bennett NC. 2005. Order Rodentia. In: Skinner JD and Chimimba CT, editors. The mammals of the southern African subregion, 3rd edition. Cape Town: Cambridge University Press; 2005. p. 77–209.
- Russo IR, Chimimba CT, Bloomer P. Bioregion heterogeneity correlates with extensive mitochondrial DNA diversity in the Namaqua rock mouse, Micaelamys namaquensis (Rodentia: Muridae) from southern Africa—evidence for a species complex. BMC Evol Biol. 2010;10:307. DOIPubMedGoogle Scholar
- Do Linh San E, Babu N, Xalu M, Le Gars S, Perquin J-C, Baxter RM, et al. A conservation assessment of Otomys unisulcatus. In: Child MF, Roxburgh L, Do Linh San E, Raimondo D, Davies-Mostert HT, editors. The red list of mammals of South Africa, Swaziland and Lesotho 2016. Pretoria, South Africa: South African National Biodiversity Institute and Endangered Wildlife Trust; 2017.
- Monadjem A, Taylor PJ, Denys C, Cotterill FPD. Rodents of sub-Saharan Africa: a biogeographic and taxonomic synthesis. Berlin: Walter de Gruyter GmbH; 2015.
- Castiglia R, Solano E, Makundi RH, Hulselmans J, Verheyen E, Colangelo P. Rapid chromosomal evolution in the mesic four-striped grass rat Rhabdomys dilectus (Rodentia, Muridae) revealed by mtDNA phylogeographic analysis. J Zool Syst Evol Res. 2011;50:165–72. DOIGoogle Scholar
- du Toit N, van Vuuren BJ, Matthee S, Matthee CA. Biome specificity of distinct genetic lineages within the four-striped mouse Rhabdomys pumilio (Rodentia: Muridae) from southern Africa with implications for taxonomy. Mol Phylogenet Evol. 2012;65:75–86. DOIPubMedGoogle Scholar
- Ganem G, Dufour C, Avenant N, Caminade P, Eiseb S, Tougard C, et al. An update on the distribution and diversification of Rhabdomys sp. (Muridae, Rodentia). J Vert Biol. 2020;69:1. DOIGoogle Scholar
- McIntosh BM, Dickinson DB, Meenehan GM, Dos Santos IS. Culex (Eumelanomyia) rubinotus Theobald as vector of Banzi, Germiston and Witwatersrand viruses. II. Infections in sentinel hamsters and wild rodents. J Med Entomol. 1976;12:641–4. DOIPubMedGoogle Scholar
- Monath TP, Newhouse VF, Kemp GE, Setzer HW, Cacciapuoti A. Lassa virus isolation from Mastomys natalensis rodents during an epidemic in Sierra Leone. Science. 1974;185:263–5. DOIPubMedGoogle Scholar
- Walker DH, Johnson KM, Lange JV, Gardner JJ, Kiley MP, McCormick JB. Experimental infection of rhesus monkeys with Lassa virus and a closely related arenavirus, Mozambique virus. J Infect Dis. 1982;146:360–8. DOIPubMedGoogle Scholar
- Paweska JT, Sewlall NH, Ksiazek TG, Blumberg LH, Hale MJ, Lipkin WI, et al.; Outbreak Control and Investigation Teams. Nosocomial outbreak of novel arenavirus infection, southern Africa. Emerg Infect Dis. 2009;15:1598–602. DOIPubMedGoogle Scholar
- Palacios G, Savji N, Hui J, Travassos da Rosa A, Popov V, Briese T, et al. Genomic and phylogenetic characterization of Merino Walk virus, a novel arenavirus isolated in South Africa. J Gen Virol. 2010;91:1315–24. DOIPubMedGoogle Scholar
- Colangelo P, Verheyen E, Leirs H, Tatard C, Denys C, Dobigny G, et al. A mitochondrial phylogeographic scenario for the most widespread African rodent, Mastomys natalensis. Biol J Linn Soc Lond. 2013;108:901–16. DOIGoogle Scholar
- Göuy de Bellocq J, Bryjová A, Martynov A, Lavrenchenko L. Dhati Welel virus, the missing mammarenavirus of the widespread Mastomys natalensis. J Vert Biol. 2020;69:20018. DOIGoogle Scholar
- Gryseels S, Baird SJE, Borremans B, Makundi R, Leirs H, Goüy de Bellocq J. When viruses don’t go viral: the importance of host phylogeographic structure in the spatial spread of arenaviruses. PLoS Pathog. 2017;13:
e1006073 . DOIPubMedGoogle Scholar - Radoshitzky SR, Buchmeier MJ, Charrel RN, Clegg JCS, Gonzalez JJ, Günther S, et al.; Ictv Report Consortium. ICTV virus taxonomy profile: Arenaviridae. J Gen Virol. 2019;100:1200–1. DOIPubMedGoogle Scholar
- Witkowski PT, Kallies R, Hoveka J, Auste B, Ithete NL, Šoltys K, et al. Novel arenavirus isolates from Namaqua rock mice, Namibia, Southern Africa. Emerg Infect Dis. 2015;21:1213–6. DOIPubMedGoogle Scholar
- Těšíková J, Krásová J, Goüy de Bellocq J. Multiple mammarenaviruses circulating in Angolan rodents. Viruses. 2021;13:982. DOIPubMedGoogle Scholar
- Edwards S, Claude J, Van Vuuren BJ, Matthee CA. Van Vuuren Bj, Matthee Ca. Evolutionary history of the Karoo bush rat, Myotomys unisulcatus (Rodentia: Muridae): disconcordance between morphology and genetics. Biol J Linn Soc Lond. 2011;102:510–26. DOIGoogle Scholar
- Ishii A, Thomas Y, Moonga L, Nakamura I, Ohnuma A, Hang’ombe BM, et al. Molecular surveillance and phylogenetic analysis of Old World arenaviruses in Zambia. J Gen Virol. 2012;93:2247–51. DOIPubMedGoogle Scholar
1Current affiliation: EduVos, Midrand, South Africa.
2Current affiliation: University of the Free State, Bloemfontein, South Africa.
3Current affiliation: University of Pretoria, Pretoria, South Africa.