Volume 27, Number 2—February 2021
Dispatch
Azithromycin-Resistant Salmonella enterica Serovar Typhi AcrB-R717Q/L, Singapore
Abstract
Global travel has led to intermittent importation of multidrug-resistant Salmonella enterica serovar Typhi into industrialized countries. We detected azithromycin-resistant Salmonella Typhi in Singapore, of which 2 isolates were likely locally acquired. Ongoing vigilance and surveillance to minimize the public health risk for this serious pathogen is needed.
In Singapore, the incidence of typhoid fever is low (0.8–1.2 cases/100,000 population annually). Most cases are imported, particularly from the South Asia subcontinent (1). First-line treatments include ampicillin, trimethoprim/sulfamethoxazole, and chloramphenicol. With increasing multidrug-resistant and fluoroquinolone-resistant infections, ceftriaxone and azithromycin are the next treatment alternatives. However, resistance to ceftriaxone or azithromycin resistance has been reported (2).
Multidrug-resistant Salmonella Typhi isolates belong to haplotype H58 (genotype 4.3.1), which is predominant in Asia and Africa (3). Resistance in genotype 4.3.1 is characterized by nonsynonymous mutations in the quinolone resistance–determining-region (QRDR) of DNA gyrase genes gyrA and gyrB, DNA topoisomerase IV genes parC and parE, and acquisitions of IncHI1 plasmids (3,4). Azithromycin-resistant Salmonella Typhi is also seen in this genotype (2).
During September 2019–April 2020, increased MICs for azithromycin were detected for 3 Salmonella Typhi isolates identified at the National University Hospital, Singapore. To characterize the molecular mechanisms of azithromycin resistance and genetic lineage in these isolates, we performed whole-genome-sequencing.
This study was approved by the National Healthcare Group Domain Specific Review Board (study no. 2020/01010). Apart from the 3 isolates tested for azithromycin resistance, an additional 21 Salmonella Typhi isolates (total 24 isolates) collected during 2016 and 2020 at the National University Hospital, a 1,200-bed tertiary hospital, were retrospectively investigated.
Genus was identified by using the Bruker MALDI Biotyper (Bruker Daltonics, https://www.bruker.com), and serotyping was performed by using slide agglutination and antiserum (Statens Serum Institute, Copenhagen, Denmark). After genus and serotype were confirmed, these isolates were submitted to the National Public Health Laboratory, Singapore, as part of the national surveillance program for Salmonella spp.
Drug susceptibility testing was routinely performed by using Vitek 2 (bioMérieux, https://www.biomerieux.com) and supplemented by using the Etest (bioMérieux) for ciprofloxacin and azithromycin. Azithromycin MICs were further confirmed by using broth microdilution. Quality control isolates used were Escherichia coli ATCC 25922, Salmonella Enteritidis ATCC 13076, Salmonella Typhimurium ATCC 14028, and Staphylococcus aureus ATCC 29213. EUCAST interpretative breakpoints were used, including for azithromycin resistance, which is based on the tentative epidemiologic cutoff value (https://eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_10.0_Breakpoint_Tables.pdf).
Whole-genome sequencing was performed by using MiSeq (Illumina, https://www.illumina.com) to generate 300-bp paired-end reads. Raw reads were assembled by using Shovill (https://github.com/tseemann/shovill). Isolates were genotyped by using the GenoTyphi tool (https://github.com/katholt/genotyphi), which separates Salmonella Typhi isolates into clades on the basis of the extended genotyping framework described by Wong et al. (4). SRST2 (5) was used to determine the presence of acquired antimicrobial drug resistance genes by using the ResFinder database (6). Chromosomal QRDR mutations in gyrA, gyrB, and parC, as well as the efflux pump AcrB (acrB-R717Q) mutations conferring resistance to azithromycin, were also investigated by using the GenoTyphi tool. PlasmidFinder (https://cge.cbs.dtu.dk/services/PlasmidFinder/) was used to detect replicons.
Salmonella Typhi CT18 (GenBank accession no. AL513382.1) was designated as the reference genome. We also downloaded all publicly available Salmonella Typhi genome sequences belonging to lineage 4.3.1 and its sublineages from the Pathogenwatch database (https://pathogen.watch) for comparison with our isolates belonging to lineage 4.3.1. Core-genome single-nucleotide polymorphisms were obtained by using snippy pipeline (https://github.com/tseemann/snippy) and then used to generate a phylogenetic tree by using FastTree (7). The resulting tree was visualized by using iTOL version 4 (8). Raw reads have been submitted to the Sequence Reads Archive under BioProject no. PRJNA660881.
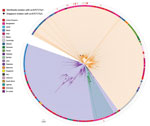
Whole-genome sequencing results showed that 15 of the 24 Salmonella Typhi isolates belonged to subclade 4.3.1 (haplotype H58), which can be further differentiated into 4.3.1.1 (4/15), 4.3.1.2 (8/15), and 4.3.1.3 (3/15) (Table). The 4.3.1 subclade is a dominant lineage disseminating from South Asia into East Africa (3). Signature mutations associated with this subclade are QRDR mutations at codon positions 83 and 87 in gyrA conferring fluoroquinolone resistance. The phylogenetic tree showed that these isolates did not form a unique group but were interspersed with isolates from countries in South Asia, particularly Bangladesh (Figure). The remaining 9/24 isolates belonged to subclades 0.0.2 (n = 1), 2.3.3 (n = 4), 3 (n = 2), 3.2.1 (n = 1), and 4.1 (n = 1).
The genomic antimicrobial drug–susceptibility profiles correlated with the phenotypic susceptibilities (Table). Six isolates harbored blaTEM-1, but none had extended-spectrum-β-lactamases or carbapenemases. All isolates were resistant to ciprofloxacin and had QRDR mutations (Table). Four of 8 isolates belonged to subclade 4.3.1.2 and had the triple QRDR mutation combination. Only isolates with dfrA7, sul1, and sul2 were phenotypically resistant to trimethoprim/sulfamethoxazole.
The 3 azithromycin-resistant isolates have not acquired macrolide-modifying enzymes, such as methylases [erm(A), erm(B), and erm(C)], esterases [ere(A) and ere(B)], or phosphotransferases [mph(A), mph(B), and mph(D)] observed in isolates belonging to the order Enterobacterales (9). There were no chromosomal alterations in the 50S ribosomal subunit proteins L4 (rlpD) and L22 (rlpV) (11). Instead, R717Q/L mutations in the efflux pump AcrB were detected. Increased MICs for azithromycin (R717Q: 32 mg/L, R717L: 16 mg/L) were observed for these isolates. Azithromycin MICs <4 mg/L were observed for all wild-type acrB isolates (Table). The AcrB-R717Q mutation was reported in azithromycin-resistant Salmonella Typhi 4.3.1.1 in Bangladesh (2) and subsequently in a Pakistan-specific 4.3.1.1 cluster (10). The mutation that emerged in Pakistan is believed to be a de novo spontaneous mutation, rather than spread of an azithromycin-resistant clone (10). AcrB-R717Q–associated azithromycin resistance has also been reported in India (11).
The AcrB-R717L mutation (isolate SLT1105) is novel in Salmonella Typhi. This mutation was described in an azithromycin-resistant Salmonella Paratyphi A isolate in Bangladesh (2). Functional analysis of the R717L mutation conferred resistance to a sensitive Salmonella Paratyphi A strain resulted in a 4-fold increase in the MIC (7 mg/L vs. 28 mg/L; p = 0.0001) (2). In Salmonella Typhi, the mutation also appears to impart azithromycin resistance (Table).
These AcrB-R717Q/L mutations were in multidrug-resistant isolates. This finding is worrisome because of the unavailability of oral antimicrobial drug treatment options and increased relapses when treated with β-lactams without intracellular-acting antimicrobial drugs.
Most case-patients had relevant travel history within 2 months before onset of symptoms, including travel to India (n = 5), Bangladesh (n = 4), Pakistan (n = 1), Myanmar (n = 1), and the Philippines (n = 1). Three cases appeared to be local transmission, of which 2 had AcrB-R717Q/L mutations. The remaining case-patient, whose isolate had the AcrB-R717Q mutation, had traveled to Bangladesh and probably acquired the infection in this country (2).
Hooda et al. (12) analyzed 49,115 Salmonella genomes and found the AcrB-R717Q/L mutation in 16 Salmonella Typhi genomes (≈0.03%). Although this number was small, the rate of acquisition of such novel mechanisms might hasten, especially with increasing use of azithromycin, such as in mass drug administration with azithromycin as a key component for the control of neglected tropical diseases (12). The proportion of isolates in our study with AcrB-717Q/L mutation (20%, 3/15) was unexpectedly higher than previously reported. The reasons for this finding are unclear. However, our study was a single-center study that had a limited number of cases.
Genotypic testing of the usual azithromycin resistance–associated genes in the order Enterobacterales cannot identify acrB mutations, and there are currently no formal breakpoints to guide phenotypic testing. An azithromycin MIC of 16 mg/L was observed for 1 isolate with the AcrB-R717L mutation. Although this azithromycin MIC was higher (<8 mg/L) than that for isolates without the mutation, this isolate is still considered wild-type. Detection of increased MICs raises suspicion for resistance requiring further confirmation. Additional data are required to correlate resistance mutations, MICs, and treatment outcomes.
Dr. Octavia is a scientific officer at the National Public Health Laboratory, Singapore. Her primary research interest is laboratory surveillance of salmonellosis.
References
- Yew FS, Goh KT, Lim YS. Epidemiology of typhoid fever in Singapore. Epidemiol Infect. 1993;110:63–70. DOIPubMedGoogle Scholar
- Hooda Y, Sajib MSI, Rahman H, Luby SP, Bondy-Denomy J, Santosham M, et al. Molecular mechanism of azithromycin resistance among typhoidal Salmonella strains in Bangladesh identified through passive pediatric surveillance. PLoS Negl Trop Dis. 2019;13:e0007868–0007868. DOIPubMedGoogle Scholar
- Wong VK, Baker S, Pickard DJ, Parkhill J, Page AJ, Feasey NA, et al. Phylogeographical analysis of the dominant multidrug-resistant H58 clade of Salmonella Typhi identifies inter- and intracontinental transmission events. Nat Genet. 2015;47:632–9. DOIPubMedGoogle Scholar
- Wong VK, Baker S, Connor TR, Pickard D, Page AJ, Dave J, et al.; International Typhoid Consortium. An extended genotyping framework for Salmonella enterica serovar Typhi, the cause of human typhoid. Nat Commun. 2016;7:12827. DOIPubMedGoogle Scholar
- Inouye M, Dashnow H, Raven L-A, Schultz MB, Pope BJ, Tomita T, et al. SRST2: Rapid genomic surveillance for public health and hospital microbiology labs. Genome Med. 2014;6:90. DOIPubMedGoogle Scholar
- Bortolaia V, Kaas RS, Ruppe E, Roberts MC, Schwarz S, Cattoir V, et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother. 2020;dkaa345.
- Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26:1641–50. DOIPubMedGoogle Scholar
- Letunic I, Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47(W1):W256–9. DOIPubMedGoogle Scholar
- Gomes C, Martínez-Puchol S, Palma N, Horna G, Ruiz-Roldán L, Pons MJ, et al. Macrolide resistance mechanisms in Enterobacteriaceae: Focus on azithromycin. Crit Rev Microbiol. 2017;43:1–30. DOIPubMedGoogle Scholar
- Iqbal J, Dehraj IF, Carey ME, Dyson ZA, Garrett D, Seidman JC, et al. A race against time: reduced azithromycin susceptibility in Salmonella enterica serovar Typhi in Pakistan. MSphere. 2020;5:e00215–20. DOIPubMedGoogle Scholar
- Katiyar A, Sharma P, Dahiya S, Singh H, Kapil A, Kaur P. Genomic profiling of antimicrobial resistance genes in clinical isolates of Salmonella Typhi from patients infected with Typhoid fever in India. Sci Rep. 2020;10:8299. DOIPubMedGoogle Scholar
- Hooda Y, Tanmoy AM, Sajib MSI, Saha S. Mass azithromycin administration: considerations in an increasingly resistant world. BMJ Glob Health. 2020;5:
e002446 . DOIPubMedGoogle Scholar
Figure
Table
Cite This ArticleOriginal Publication Date: January 20, 2021
Table of Contents – Volume 27, Number 2—February 2021
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Sophie Octavia, National Public Health Laboratory, National Centre for Infectious Diseases, 16 Jalan Tan Tock Seng, Singapore 308442
Top