Volume 27, Number 4—April 2021
Research
Epidemiologic and Genomic Reidentification of Yaws, Liberia
Figure 4
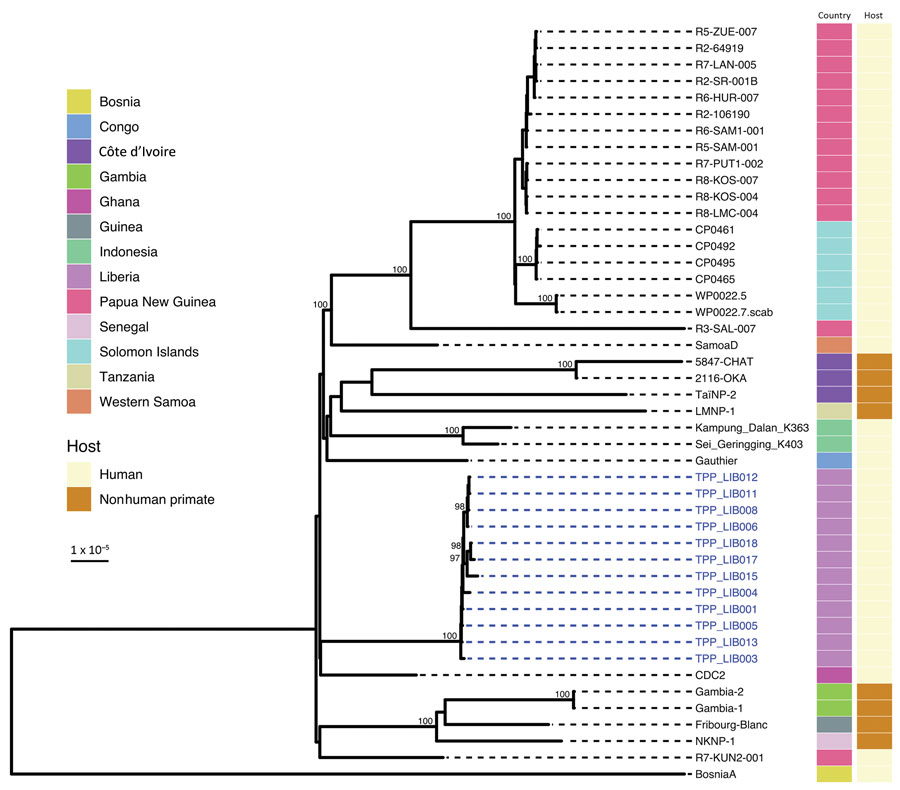
Figure 4. Global context of whole Treponema pallidum subspecies pertenue (TPE) genomes from Liberia. Liberia genomes form a monophyletic clade, genetically distant from publicly available genomes including 3 isolated from nonhuman primates in nearby Taï National Park (Côte d’Ivoire). Plot shows a maximum-likelihood phylogeny of 12 Liberia genomes contextualized with 34 published global genomes, scaled by substitutions per site. Ultra-fast bootstrap values >95% are indicated on the tree. Colored tracks show country of sampling and original host organism. Novel Liberia genomes from this study are indicated with blue labels.
1These senior authors contributed equally to this article.
Page created: January 13, 2021
Page updated: March 18, 2021
Page reviewed: March 18, 2021
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.