Volume 27, Number 4—April 2021
Dispatch
Rare Norovirus GIV Foodborne Outbreak, Wisconsin, USA
Figure 2
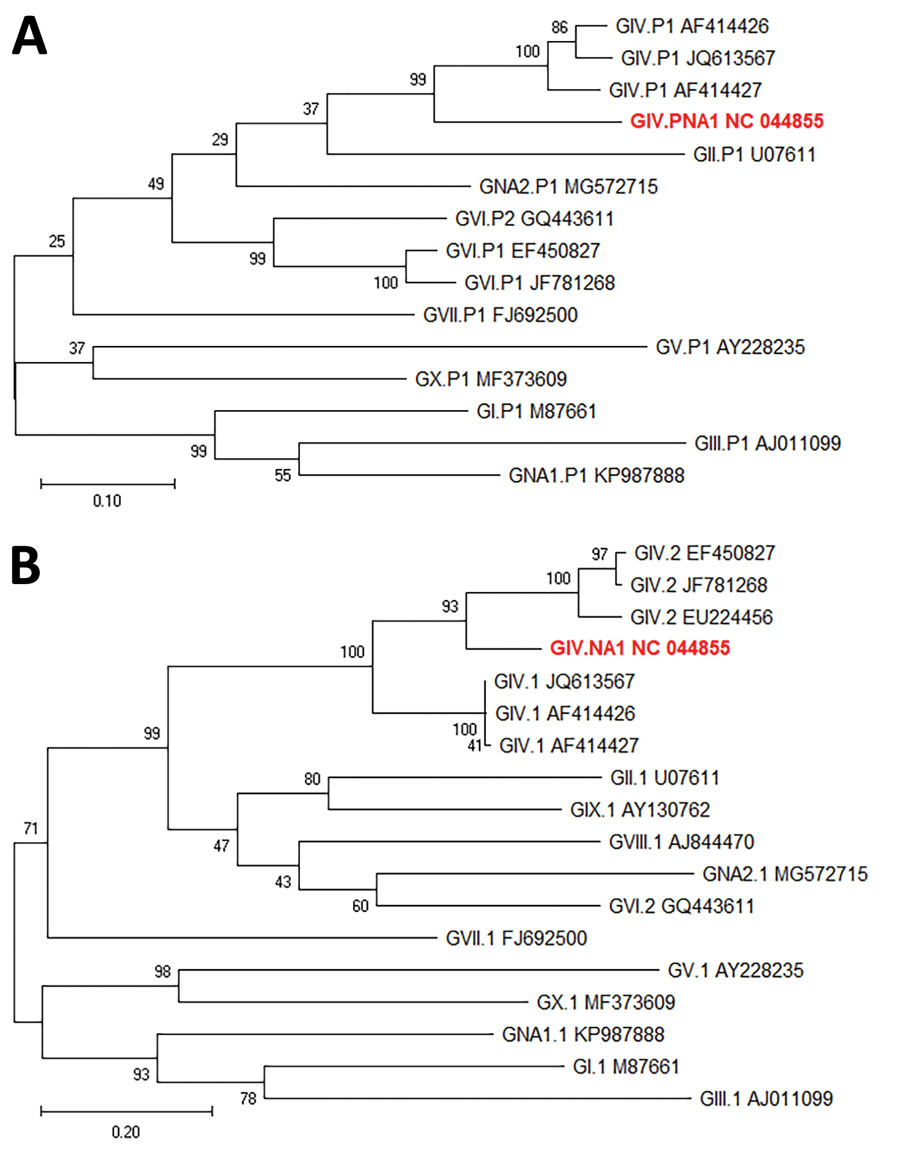
Figure 2. Maximum-likelihood phylogenetic analysis of rare norovirus GIV isolated during foodborne outbreak, Wisconsin, USA (red text), and reference strains. A) Partial polymerase gene (762 nt); B) complete capsid (VP1) gene (554 aa). Bootstrap support for 500 replicates is indicated on branches. For polymerase analysis, evolutionary distances were inferred by the Tamura-Nei model. For VP1 analysis, evolutionary distances were inferred by using the Jones-Taylor-Thornton matrix-based model. Reference strains are represented by type and GenBank accession number. Scale bar in panel A indicates nucleotide substitutions per site, and scale bar in panel B indicates amino acid substitutions per site.
Page created: January 19, 2021
Page updated: March 18, 2021
Page reviewed: March 18, 2021
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.