Volume 27, Number 8—August 2021
Research
Spatial, Ecologic, and Clinical Epidemiology of Community-Onset, Ceftriaxone-Resistant Enterobacteriaceae, Cook County, Illinois, USA
Cite This Article
Citation for Media
Abstract
We performed a spatial and mixed ecologic study of community-onset Enterobacteriaceae isolates collected from a public healthcare system in Cook County, Illinois, USA. Individual-level data were collected from the electronic medical record and census tract–level data from the US Census Bureau. Associations between individual- and population-level characteristics and presence of ceftriaxone resistance were determined by logistic regression analysis. Spatial analysis confirmed nonrandom distribution of ceftriaxone resistance across census tracts, which was associated with higher percentages of Hispanic, foreign-born, and uninsured residents. Individual-level analysis showed that ceftriaxone resistance was associated with male sex, an age range of 35–85 years, race or ethnicity other than non-Hispanic Black, inpatient encounter, and percentage of foreign-born residents in the census tract of isolate provenance. Our findings suggest that the likelihood of community-onset ceftriaxone resistance in Enterobacteriaceae is influenced by geographic and population-level variables. The development of effective mitigation strategies might depend on better accounting for these factors.
The continuous rise of infections secondary to extended-spectrum beta-lactamase (ESBL)–producing Enterobacteriaceae in the United States is a complex public health problem and considered a serious threat by the Centers for Disease Control and Prevention (1). Recently, the incidence of infections caused by ESBL producers in the United States was noted to have increased by 53.3% during 2019–2017, driven largely by a surge in community-onset cases (2). Globally, a similar trend has been described, and developing countries bear a disproportionate burden of infections secondary to these drug-resistant pathogens (3–5). The steady increases in rates of infections caused by ESBL-producing Escherichia coli and Klebsiella pneumoniae persist despite antimicrobial stewardship and infection control efforts (6,7).
Initially confined to the healthcare environment, infections caused by ESBL-producing Enterobacteriaceae among patients without previous healthcare exposure have been described since the mid-2000s (8,9). This epidemiologic shift has been largely attributed to the emergence of the CTX-M–producing E. coli sequence type (ST) 131 clone, which expanded rapidly throughout the United States and remains the most prevalent ESBL-producing E. coli clone in the community (10). In addition to higher virulence and transmissibility of the E. coli ST131 clone, its therapeutic management is particularly challenging because of its associated resistance to commonly used oral antimicrobial drugs such as quinolones and trimethoprim/sulfamethoxazole (6,10).
From an epidemiologic standpoint, multiple transmission pathways for community-onset ESBL-producing Enterobacteriaceae have been proposed. Potential sources of acquisition outside of healthcare environments include gastrointestinal colonization after international travel (11,12) and transmission among household members (7,13). In addition, ESBL-producing Enterobacteriaceae have been isolated from foodstuffs (14,15), livestock (14), and waterways (16,17), all of which have been posited as potential sources for human colonization and subsequent infection. A better understanding of the epidemiology of community-onset infections caused by ESBL-producing bacteria across geographic areas can help identify areas with higher disease burden and suggest pathways of transmission and mitigation strategies that are potentially unique to each region. Spatial and ecologic analyses help to address the influence of geography and population-level variables on disease distribution in a given region.
We conducted an epidemiologic analysis of the distribution of community-onset, ceftriaxone-resistant (CTX-R) Enterobacteriaceae from a single healthcare system in Cook County, Illinois, USA. We hypothesized that population-level characteristics are contributing factors for the presence of CTX-R Enterobacteriaceae in a geographic area and at the individual level.
Cook County Health (CCH) is a large safety-net healthcare system in Chicago and suburban Cook County, Illinois. It consists of a 450-bed teaching hospital near downtown Chicago, a small community hospital in the South Side of Chicago, a small hospital and clinic for the treatment of detainees in the Cook County jail, and 13 community clinics distributed throughout Cook County. In 2018, CCH cared for 205,322 persons, most of whom self-identified as non-Hispanic Black (49.1%) or Hispanic (32.7%). Through electronic queries, we identified all culture isolates of the commonest Enterobacteriaceae species collected at CCH: E. coli, K. pneumoniae, Enterobacter cloacae, Proteus mirabilis, Enterobacter aerogenes, and Klebsiella oxytoca collected from Cook County residents during January 1, 2016–December 31, 2018. We determined antimicrobial susceptibilities by using the MicroScan Gram-negative panel (Beckman Coulter, https://www.beckmancoulter.com) and interpreted results by using Clinical and Laboratory Standards Institute breakpoints (18). We obtained antimicrobial susceptibilities retrospectively and did not retain any isolates for further analysis. We excluded isolates collected from persons <18 years of age, surveillance isolates, isolates with intermediate susceptibility to ceftriaxone or intermediate susceptibility or resistance to carbapenems, and duplicate isolates (defined as isolates from the same persons, of the same species, and collected within 30 days). To select for community-onset isolates, we included only isolates collected in the ambulatory clinic or emergency department (ED) setting and those collected during the first 2 days of hospitalization.
Demographic characteristics, collected from the electronic medical record (EMR), were patient sex and age and self-identified race and ethnicity, categorized as non-Hispanic Black, non-Hispanic White, Hispanic, or other. We classified encounter types as outpatient (ambulatory clinic), ED, or inpatient. Census-tract variables for Cook County were obtained from the 2017 US Census Bureau American Community Survey 5-year estimates (19). We extracted census tract data on race and ethnicity, immigration status (US-born or foreign-born), deprivation (households below poverty level and uninsured status), and overcrowding (>1.5 occupants per room).
Spatial Analysis
Cook County, which includes the city of Chicago, contains 1,319 land census tracts and has an estimated population of 5,149,580 residents (19). We used ArcGIS version 10.4.1 (ESRI, https://www.esri.com) to geocode isolates to their census tract of provenance by using residential addresses available in the EMR. We calculated and mapped the percentage of CTX-R isolates in each census tract (i.e., the number of CTX-R isolates divided by the number of all isolates multiplied by 100). To minimize imprecision of CTX-R percentages in census tracts with low number of isolates, we excluded from the spatial analysis census tracts that had <3 isolates collected during the study period. We used spatial autocorrelation analysis (Moran I) to identify whether Enterobacteriaceae CTX-R percentages were distributed at random or clustered in census tracts across Cook County. Similarly, we conducted spatial autocorrelation analysis on CTX-R percentage distribution of E. coli isolates alone.
Ecologic Analysis
After excluding census tracts with <3 isolates, we categorized the remaining census tracts on the basis of the presence or absence of a CTX-R isolate. We evaluated the relationship between each population-level variable and the presence of >1 CTX-R isolates in a census tract by using bivariate logistic regression, summarized by odds ratios (ORs) and corresponding 95% CIs. We conducted a similar analysis for E. coli isolates alone.
Individual Risk Analysis
We categorized individual Enterobacteriaceae isolates on the basis of the identification of ceftriaxone resistance in the susceptibility panel. We included all isolates in the analysis of individual risk. The variables of interest were the individual demographic variables collected from the EMR and the type of clinical encounter. In addition, we included an ecologic variable, the percentage of foreign-born population in the census tract of residency. We evaluated the relationship between each variable and identification of ceftriaxone resistance in an individual isolate by using bivariate logistic regression, summarized by ORs and corresponding 95% CIs. We conducted all statistical analyses by using Stata version 14.2 (StataCorp, https://www.stata.com).
We collected 12,892 Enterobacteriaceae isolates at CCH during the study period, 10,891 of which met the inclusion criteria and were included in the dataset. We summarized the demographic and clinical characteristics of the patients from whom Enterobacteriaceae isolates were collected (Table 1). Most isolates were collected from women (7,853 [72.1%]), were from urine specimens (9,315 [85.5%]), were collected in ambulatory clinics (5,889 [54.1%]), or were identified as E. coli (7,977 [73.2%]). A total of 1,035 (9.5%) Enterobacteriaceae (817 [10.2%] E. coli isolates) were resistant to ceftriaxone. We observed no notable trends in ceftriaxone resistance across study years.
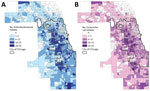
In the 1,319 land census tracts in Cook County, we collected Enterobacteriaceae isolates from residents of 1,131 (85.8%) and E. coli alone from residents of 1,085 (82.3%). The mean number of such isolates per census tract was 9.6 (SD + 9.28, range 1–92), and the mean number of E. coli isolates obtained per census tract was 7.4 (SD + 7.16, range 1–62). We plotted choropleth maps depicting the geographic distribution of all Enterobacteriaceae isolates and E. coli isolates alone (Figure 1). Among census tracts from which >1 isolate was obtained, CTX-R Enterobacteriaceae isolates were identified in 500 (44.2%), and most CTX-R isolates (561 [54.2%]) came from only 125 (11%) census tracts. In the case of CTX-R E. coli isolates, 424 (39.1%) of the 1,085 census tracts had a CTX-R E. coli isolate reported during the study period, and only 93 (8.6%) census tracts accounted for 406 (49.7%) of all CTX-R E. coli isolates.
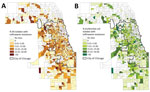
A total of 886 census tracts had >3 Enterobacteriaceae isolates collected during the study period and were included in the spatial and ecologic analyses. The mean CTX-R percentage among these census tracts was 8.7%. Autocorrelation analysis (Moran I) indicated that CTX-R percentages among all isolates were not distributed randomly across Cook County census tracts (index 0.02, p<0.01). A total of 776 census tracts had >3 E. coli isolates collected during the study period and were included in the spatial and ecologic analysis of E. coli isolates. The average CTX-R percentage of E. coli isolates among census tracts was 9.6%. Autocorrelation analysis (Moran I) of CTX-R percentages among E. coli isolates also found a nonrandom distribution among census tracts (index 0.03, p<0.01). We mapped the geographic distribution of CTX-R percentages for all Enterobacteriaceae and for E. coli isolates alone (Figure 2).
We identified census tract–level characteristics reported in the 2017 American Community Survey of residents of the 886 census tracts that accounted for >3 Enterobacteriaceae isolates and compared census tracts with ceftriaxone resistance (461 [52.1%] of census tracts, mean 15.5 isolates/census tract) and without (425 [47.9%] of census tracts, mean 8.03 isolates/census tract). Bivariate analysis found that the presence of CTX-R isolates was negatively associated with census-tract percentages of non-Hispanic White and non-Hispanic Black populations, and positively associated with census-tract percentages of Hispanic, foreign-born, and uninsured residents. We observed no statistical associations between the outcome and percentages of households with incomes below the federal poverty level or with overcrowding (Table 2). Census tract-level characteristics were moderately correlated (r = −0.78 to 0.69).
Among the 776 census tracts with >3 E. coli isolates, 395 (50.9%) had no CTX-R isolates and 381 (49.1%) had >1 resistant E. coli isolate collected during the study period, with an average CTX-R percentage of 19.4%. Bivariate analysis showed a negative association between presence of CTX-R E. coli isolates in census tracts and percentage of non-Hispanic Black population. Conversely, the odds of ceftriaxone resistance in an E.coli isolates was positively associated with the percentage of Hispanic, foreign-born, and uninsured residents and with residential overcrowding (Table 3).
All 10,891 Enterobacteriaceae isolates (1,035 [9.5%] of which were CTX-R) were included in the individual risk analysis of patients from whom CTX-R and CTX-susceptible Enterobacteriaceae were recovered (Table 4). In the bivariate logistic regression analysis, male sex, an age range of 35–85 years, race and ethnicity other than non-Hispanic Black, and inpatient encounter were found to be associated with a higher likelihood of ceftriaxone resistance in a clinical isolate. Similarly, higher odds for the outcome were associated with the percentage of foreign-born residents in the census tract of isolate provenance.
Our study has 4 main findings. First, compared with patients from whom CTX-susceptible community-onset Enterobacteriaceae isolates were collected, patients with CTX-R isolates more often were male, were 35–85 years of age, had self-identified race and ethnicity other than non-Hispanic Black, were hospitalized rather than discharged from the ED or seen in clinic, and resided in Cook County census tracts with higher proportions of foreign-born residents. Second, most patients with CTX-R isolates resided in a relatively small number of census tracts, with only 11% of Enterobacteriaceae isolate–generating census tracts accounting for 54.2% of CTX-R isolates and 93 (8.6%) of E. coli isolate–generating census tracts accounting for 49.7% of all CTX-R E. coli isolates. Third, spatial analysis supported the nonrandom distribution of Cook County census tracts generating higher proportions of ceftriaxone resistance among Enterobacteriaceae and E. coli isolates. Fourth, the population-level characteristics of census tracts from which isolates of CTX-R Enterobacteriaceae and E. coli were obtained differed from residents of census tracts yielding susceptible isolates exclusively, with the percentage of Hispanic residents, foreign-born, and uninsured population being positively associated with the presence of CTX-R isolates on analysis in both cohorts.
Similar to our findings, spatial studies conducted abroad of drug-resistant Enterobacteriaceae have shown nonrandom spatial distribution of antimicrobial-resistant Enterobacteriaceae in large urban areas. A study from São Paulo, Brazil (20), identified hotspot clusters of ciprofloxacin-resistant E.coli isolates that were associated with population-level ciprofloxacin usage. A study from Japan (21) also showed clustering of levofloxacin-resistant E. coli isolates in the western part of the country, also associated with population-level quinolone usage. In Chicago, residence in the northwest and southern region of Chicago (and adjacent suburbs) was independently associated with increased likelihood of infection by CTX-M-9 Enterobacteriaceae isolates in children (22).
Our individual-level analysis showing that ceftriaxone resistance was associated with increasing age and male sex is consistent with data reported elsewhere (8,23) and might reflect unmeasured associated underlying conditions, especially those involving the genitourinary tract (8,24) and antibiotic exposures (8,25–27). Unmeasured underlying conditions and associated antibiotic exposure could also account for the strong association between ceftriaxone resistance and the need for hospitalization, although the increased virulence observed in circulating ESBL-producing clones (28) could account for this finding.
The associations between self-reported Hispanic ethnicity and CTX-R Enterobacteriaceae and E. coli identified in the individual-level analysis and ecologic analyses merit further scrutiny. First, the correlation of Hispanic ethnicity and foreign-born status at a population level (r = 0.69) suggests that these 2 communities are highly interrelated; indeed, ≈45.6% of foreign-born persons in Cook County are noted to have emigrated from Latin America (19). Therefore, patients who self-identified as Hispanics also might have been foreign-born and might have become colonized by resistant organisms before emigration from or during travel to Latin American countries, some of which have reported high prevalence of ESBL-producing Enterobacteriaceae (5,29,30). This same pathway could explain the similar association between the proportion of foreign-born population in a census tract and likelihood of ceftriaxone-resistance in the ecologic and individual-level analyses. In addition, a sizable proportion of non–US-born Cook County residents emigrated from countries in Asia (27.3%) and fewer emigrated from Africa (3.2%) (19), continents with variable but often high prevalence of drug-resistant Enterobacteriaceae (3,4) (We did not include other racial and ethnic population-level characteristics in our individual-level analysis because of multicollinearity with individual-level race and ethnicity). Second, Hispanics residing in the United States have been reported to use antibiotics without prescription more frequently than other racial and ethnic groups (31). Third, we cannot discount that proximity of Hispanic communities, foreign-born communities, or both to environmental sources, such as contaminated waterways, might be an important added risk factor for colonization or infection by drug-resistant Enterobacteriaceae in these areas.
Although in our ecologic analysis the percentage of households beneath the poverty line was not significantly different between census tracts from which CTX-R Enterobacteriaceae or E. coli isolates were and were not generated, observed associations between the percentage of uninsured residents and the presence in census tracts of CTX-R isolates suggest that census tract-level deprivation might predispose to antimicrobial-resistant infections. In the analysis limited to E. coli isolates, overcrowding percentages were also associated with antimicrobial-resistant infections, suggesting possible household-level transmission. A recently published study by Otter et al. from London (27) identified associations between community-level variables, individual-level variables, and likelihood of ESBL rectal colonization among patients admitted to the hospital. In their analysis, only recent overseas travel, recent antimicrobial use, and community-level overcrowding rates were associated with ESBL rectal carriage, whereas individual- and community-level race, ethnicity, and immigration characteristics were not. The paucity of spatial and ecologic studies of antimicrobial-resistant Enterobacteriaceae in the United States makes it difficult to establish whether our results are representative of the urban epidemiology of these organisms in the country. Although not directly comparable because of a difference in outcomes, the discrepancy of our findings and those reported by Otter et al. (27) suggest that the effect of population-level variables might remain distinct in different geographic areas.
Our findings are limited by the fact that our isolates were obtained in a single healthcare system. As a safety-net healthcare system, CCH is likely to be subject to geographic bias already because our patients do not come equitably from all census tracts in Cook County. The paucity of isolates from Cook County communities that do not obtain services from our healthcare system limits the generalizability of our findings regionally. We were unable to gather data regarding risk factors for healthcare-associated infections (such as recent hospitalization) and recent antimicrobial use, both important limitations. In addition, the relatively small sample size and high correlation between population-level factors made meaningful multivariable analysis infeasible. We were unable to perform genomic analysis of CTX-R organisms, which would have enabled us to evaluate the relatedness of isolates and make stronger inferences about whether spatial clustering was related to a point source or interpersonal transmission. Finally, the limited number of clinical and population-level variables included in the individual risk analysis prevents definite conclusions regarding individual risk for CTX-R infection among our patients. Indeed, concurrent assessment of other well-known individual risk factors, such as recent travel or antimicrobial use, could alter the effect size of ecologic variables. Nevertheless, our findings corroborate previous investigations that have identified important community-level variation in CTX-R infection risk in association with geographic (20–22), demographic (7,23–25), and population-level variables (27). Developing effective mitigation strategies, such as focusing antimicrobial stewardship efforts on affected areas, including residence as a risk factor in treatment-decision algorithms, or identifying and eradicating local environmental sources of drug-resistant pathogens, could well depend on improved understanding of these dynamics.
Dr. Sardá is an infectious disease attending at Cook County Health, and an assistant professor of medicine at Rush Medical College in Chicago, IL. Her research interests are in antimicrobial resistance, spatial and ecological determinants of health, and the applications of machine learning in epidemiology.
References
- CDC. Antibiotic resistance threats in the United States, 2019 [cited 2020 Oct 1]. https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf
- Jernigan JA, Hatfield KM, Wolford H, Nelson RE, Olubajo B, Reddy SC, et al. Multidrug-resistant bacterial infections in U.S. hospitalized patients, 2012–2017. N Engl J Med. 2020;382:1309–19. DOIPubMedGoogle Scholar
- Flokas ME, Karanika S, Alevizakos M, Mylonakis E. Prevalence of ESBL-producing Enterobacteriaceae in pediatric bloodstream infections: a systematic review and meta-analysis. PLoS One. 2017;12:
e0171216 . DOIPubMedGoogle Scholar - Toy T, Pak GD, Duc TP, Campbell JI, El Tayeb MA, Von Kalckreuth V, et al. Multicountry distribution and characterization of extended-spectrum β-lactamase-associated Gram-negative bacteria from bloodstream infections in sub-Saharan Africa. Clin Infect Dis. 2019;69(Suppl 6):S449–58. DOIPubMedGoogle Scholar
- Villegas MV, Kattan JN, Quinteros MG, Casellas JM. Prevalence of extended-spectrum beta-lactamases in South America. Clin Microbiol Infect. 2008;14(Suppl 1):154–8. DOIPubMedGoogle Scholar
- Lob SH, Nicolle LE, Hoban DJ, Kazmierczak KM, Badal RE, Sahm DF. Susceptibility patterns and ESBL rates of Escherichia coli from urinary tract infections in Canada and the United States, SMART 2010-2014. Diagn Microbiol Infect Dis. 2016;85:459–65. DOIPubMedGoogle Scholar
- Hilty M, Betsch BY, Bögli-Stuber K, Heiniger N, Stadler M, Küffer M, et al. Transmission dynamics of extended-spectrum β-lactamase-producing Enterobacteriaceae in the tertiary care hospital and the household setting. Clin Infect Dis. 2012;55:967–75. DOIPubMedGoogle Scholar
- Rodríguez-Baño J, Navarro MD, Romero L, Martínez-Martínez L, Muniain MA, Perea EJ, et al. Epidemiology and clinical features of infections caused by extended-spectrum beta-lactamase-producing Escherichia coli in nonhospitalized patients. J Clin Microbiol. 2004;42:1089–94. DOIPubMedGoogle Scholar
- Pitout JD, Gregson DB, Church DL, Elsayed S, Laupland KB. Community-wide outbreaks of clonally related CTX-M-14 beta-lactamase-producing Escherichia coli strains in the Calgary health region. J Clin Microbiol. 2005;43:2844–9. DOIPubMedGoogle Scholar
- Doi Y, Park YS, Rivera JI, Adams-Haduch JM, Hingwe A, Sordillo EM, et al. Community-associated extended-spectrum β-lactamase-producing Escherichia coli infection in the United States. Clin Infect Dis. 2013;56:641–8. DOIPubMedGoogle Scholar
- Arcilla MS, van Hattem JM, Haverkate MR, Bootsma MCJ, van Genderen PJJ, Goorhuis A, et al. Import and spread of extended-spectrum β-lactamase-producing Enterobacteriaceae by international travellers (COMBAT study): a prospective, multicentre cohort study. Lancet Infect Dis. 2017;17:78–85. DOIPubMedGoogle Scholar
- van Duijkeren E, Wielders CCH, Dierikx CM, van Hoek AHAM, Hengeveld P, Veenman C, et al. Long-term carriage of extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae in the general population in the Netherlands. Clin Infect Dis. 2018;66:1368–76. DOIPubMedGoogle Scholar
- Martischang R, Riccio ME, Abbas M, Stewardson AJ, Kluytmans JAJW, Harbarth S. Household carriage and acquisition of extended-spectrum β-lactamase-producing Enterobacteriaceae: A systematic review. Infect Control Hosp Epidemiol. 2020;41:286–94. DOIPubMedGoogle Scholar
- Day MJ, Hopkins KL, Wareham DW, Toleman MA, Elviss N, Randall L, et al. Extended-spectrum β-lactamase-producing Escherichia coli in human-derived and foodchain-derived samples from England, Wales, and Scotland: an epidemiological surveillance and typing study. Lancet Infect Dis. 2019;19:1325–35. DOIPubMedGoogle Scholar
- Johnson JR, Sannes MR, Croy C, Johnston B, Clabots C, Kuskowski MA, et al. Antimicrobial drug-resistant Escherichia coli from humans and poultry products, Minnesota and Wisconsin, 2002-2004. Emerg Infect Dis. 2007;13:838–46. DOIPubMedGoogle Scholar
- Fuentes MD, Gutierrez S, Sahagun D, Gomez J, Mendoza J, Ellis CC, et al. Assessment of antibiotic levels, multi-drug resistant bacteria and genetic biomarkers in the waters of the Rio Grande River between the United States–Mexico border. J Health Pollut. 2019;9:
190912 . DOIPubMedGoogle Scholar - Jørgensen SB, Søraas AV, Arnesen LS, Leegaard TM, Sundsfjord A, Jenum PA. A comparison of extended spectrum β-lactamase producing Escherichia coli from clinical, recreational water and wastewater samples associated in time and location. PLoS One. 2017;12:
e0186576 . DOIPubMedGoogle Scholar - Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing, 30th edition (M100). Wayne (PA): The Institute; 2020.
- US Census Bureau. American Community Survey 5-year estimates. 2017 [cited 2020 Oct 1]. https://data.census.gov
- Kiffer CR, Camargo EC, Shimakura SE, Ribeiro PJ Jr, Bailey TC, Pignatari AC, et al. A spatial approach for the epidemiology of antibiotic use and resistance in community-based studies: the emergence of urban clusters of Escherichia coli quinolone resistance in Sao Paulo, Brasil. Int J Health Geogr. 2011;10:17. DOIPubMedGoogle Scholar
- Terahara F, Nishiura H. Fluoroquinolone consumption and Escherichia coli resistance in Japan: an ecological study. BMC Public Health. 2019;19:426. DOIPubMedGoogle Scholar
- Logan LK, Medernach RL, Domitrovic TN, Rispens JR, Hujer AM, Qureshi NK, et al. The clinical and molecular epidemiology of CTX-M-9 group producing Enterobacteriaceae infections in children. Infect Dis Ther. 2019;8:243–54. DOIPubMedGoogle Scholar
- Ho PL, Chu YP, Lo WU, Chow KH, Law PY, Tse CW, et al. High prevalence of Escherichia coli sequence type 131 among antimicrobial-resistant E. coli isolates from geriatric patients. J Med Microbiol. 2015;64:243–7. DOIPubMedGoogle Scholar
- Calbo E, Romaní V, Xercavins M, Gómez L, Vidal CG, Quintana S, et al. Risk factors for community-onset urinary tract infections due to Escherichia coli harbouring extended-spectrum beta-lactamases. J Antimicrob Chemother. 2006;57:780–3. DOIPubMedGoogle Scholar
- Rodríguez-Baño J, Picón E, Gijón P, Hernández JR, Ruíz M, Peña C, et al.; Spanish Network for Research in Infectious Diseases (REIPI). Community-onset bacteremia due to extended-spectrum beta-lactamase-producing Escherichia coli: risk factors and prognosis. Clin Infect Dis. 2010;50:40–8. DOIPubMedGoogle Scholar
- Goodman KE, Lessler J, Cosgrove SE, Harris AD, Lautenbach E, Han JH, et al.; Antibacterial Resistance Leadership Group. A clinical decision tree to predict whether a bacteremic patient is infected with an extended-spectrum β-lactamase-producing organism. Clin Infect Dis. 2016;63:896–903. DOIPubMedGoogle Scholar
- Otter JA, Natale A, Batra R, Tosas Auguet O, Dyakova E, Goldenberg SD, et al. Individual- and community-level risk factors for ESBL Enterobacteriaceae colonization identified by universal admission screening in London. Clin Microbiol Infect. 2019;25:1259–65. DOIPubMedGoogle Scholar
- Rottier WC, Ammerlaan HS, Bonten MJ. Effects of confounders and intermediates on the association of bacteraemia caused by extended-spectrum β-lactamase-producing Enterobacteriaceae and patient outcome: a meta-analysis. J Antimicrob Chemother. 2012;67:1311–20. DOIPubMedGoogle Scholar
- García C, Horna G, Linares E, Ramírez R, Tapia E, Velásquez J, et al. Antimicrobial drug resistance in Peru. Emerg Infect Dis. 2012;18:520–1. DOIPubMedGoogle Scholar
- Winokur PL, Canton R, Casellas JM, Legakis N. Variations in the prevalence of strains expressing an extended-spectrum beta-lactamase phenotype and characterization of isolates from Europe, the Americas, and the Western Pacific region. Clin Infect Dis. 2001;32(Suppl 2):S94–103. DOIPubMedGoogle Scholar
- Mainous AG III, Cheng AY, Garr RC, Tilley BC, Everett CJ, McKee MD. Nonprescribed antimicrobial drugs in Latino community, South Carolina. Emerg Infect Dis. 2005;11:883–8. DOIPubMedGoogle Scholar
Figures
Tables
Cite This ArticleOriginal Publication Date: July 13, 2021
Table of Contents – Volume 27, Number 8—August 2021
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Vanessa Sardá, Cook County Health, 1900 W Polk St, Ste 643, Chicago, IL, 60611, USA
Top