Emergence and Evolutionary Response of Vibrio cholerae to Novel Bacteriophage, Democratic Republic of the Congo1
Meer T. Alam
2, Carla Mavian
2
, Taylor K. Paisie, Massimiliano S. Tagliamonte, Melanie N. Cash, Angus Angermeyer, Kimberley D. Seed, Andrew Camilli, Felicien Masanga Maisha, R. Kabangwa Kakongo Senga, Marco Salemi, J. Glenn Morris, and Afsar Ali

Author affiliations: University of Florida Emerging Pathogens Institute, Gainesville, Florida, USA (M.T. Alam, C. Mavian, T.K. Paisie, M.S. Tagliamonte, M.N. Cash, F.M. Maisha, M. Salemi, J.G. Morris, Jr., A. Ali); University of Florida College of Medicine, Gainesville (C. Mavian, T.K. Paisie, M.S. Tagliamonte, M.N. Cash, M. Salemi, J.G. Morris, Jr.); University of California, Berkeley, California, USA (A. Angermeyer, K.D. Seed); Chan Zuckerberg Biohub, San Francisco, California, USA (K.D. Seed); Tufts University School of Medicine, Boston, Massachusetts, USA (A. Camilli); Appui Medical Integre aux Activites de Laboratoire (AMI-LABO), Goma, Democratic Republic of the Congo (R.K.K. Senga); University of Goma, Goma (R.K.K. Senga); University of Florida College of Public Health and Health Professions, Gainesville (M.T. Alam, A. Ali)
Main Article
Figure 2
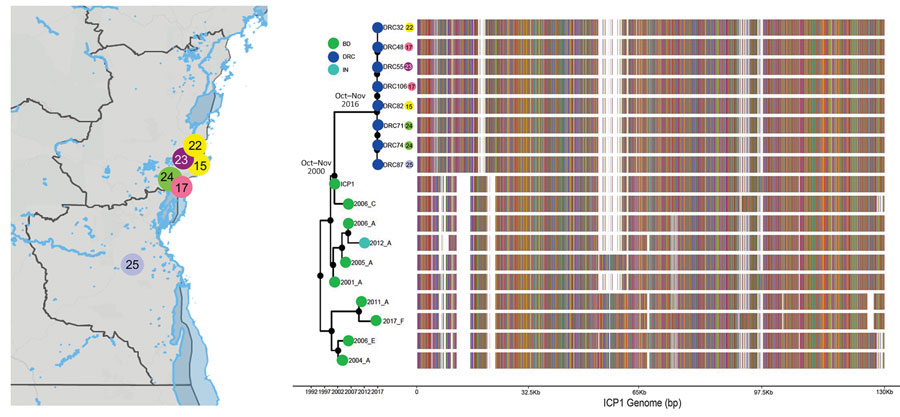
Figure 2. Bayesian inference of the phylogenetic relationship between phages and mutation patterns in the Democratic Republic of the Congo and ICP1 (Bangladesh cholera phage 1) patterns from Asia. Sampling locations of ICP1 strains from DRC are shown on the map. Each sampling location is coded by color and number, also indicated at the tip of the maximum clade credibility tree (exact locations in Appendix Table 2). The tree branches are scaled in time, and the circle tip points are colored by the location of origin, as indicated in the key. Circles in internal node indicate posterior probability support >0.9. To the right of the MCC tree, the genomic composition of each isolate is displayed: red, adenine; green, cytosine; yellow, guanine; and blue, thymine. White spaces indicate gaps at that location in the genome.
Main Article
Page created: October 18, 2022
Page updated: November 21, 2022
Page reviewed: November 21, 2022
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
