Volume 28, Number 6—June 2022
Research
Retrospective Genomic Characterization of a 2017 Dengue Virus Outbreak, Burkina Faso
Figure 1
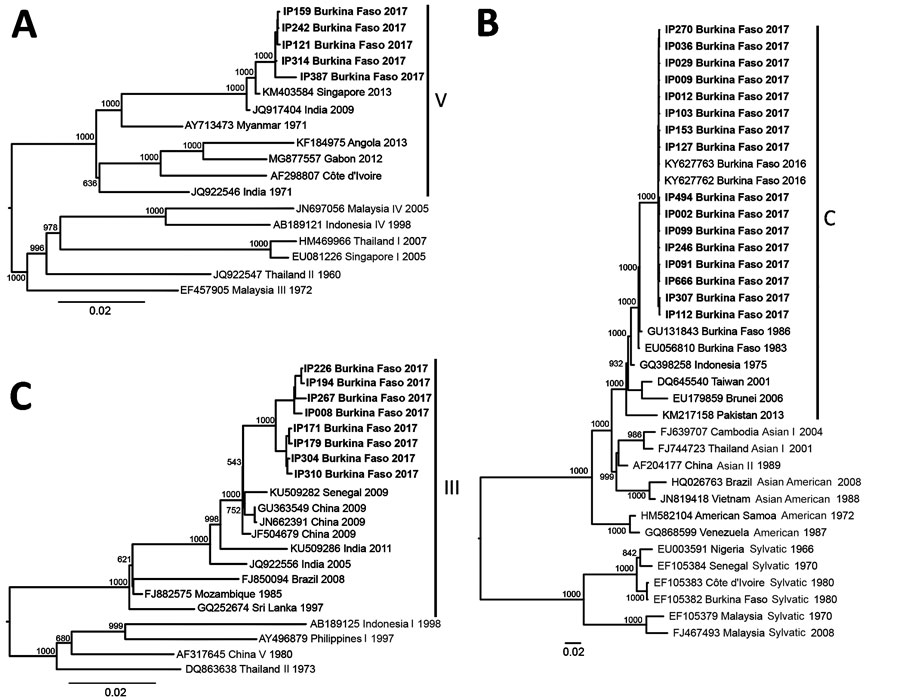
Figure 1. Phylogenetic trees of dengue virus (DENV) serotypes 1 (A), 2 (B), and 3 (C), inferred from an alignment of the 2017 Burkina Faso dengue virus outbreak genomes (boldface) and all other complete genomes from US National Institutes of Health National Institute of Allergy and Infectious Diseases Virus Pathogen Database and Analysis Resource (http://www.viprbrc.org) and pruned to representative genotypes. The Burkina Faso genomes were DENV-1 genotype V, DENV-2 genotype Cosmopolitan, and DENV-3 genotype III. GenBank accession numbers are provided for reference genomes.
1These first authors contributed equally to this article.
Page created: March 18, 2022
Page updated: May 22, 2022
Page reviewed: May 22, 2022
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.