Posttransfusion Sepsis Attributable to Bacterial Contamination in Platelet Collection Set Manufacturing Facility, United States
Ian Kracalik, Alyssa G. Kent, Carlos H. Villa, Paige Gable, Pallavi Annambhotla, Gillian McAllister, Deborah Yokoe, Charles R. Langelier, Kelly Oakeson, Judith Noble-Wang, Orieji Illoh, Alison Laufer Halpin, Anne F. Eder, and Sridhar V. Basavaraju

Author affiliations: Centers for Disease Control and Prevention, Atlanta, Georgia, USA (I. Kracalik, A.G. Kent, P. Gable, P. Annambhotla, G. McAllister, J. Noble-Wang, A.L. Halpin, S.V. Basavaraju); Food and Drug Administration, Silver Spring, Maryland, USA (C.H. Villa, O. Illoh, A.F. Eder); University of California San Francisco School of Medicine, San Francisco, California, USA (D. Yokoe, C.R. Langelier); Utah Department of Health, Salt Lake City, Utah, USA (K. Oakeson)
Main Article
Figure 5
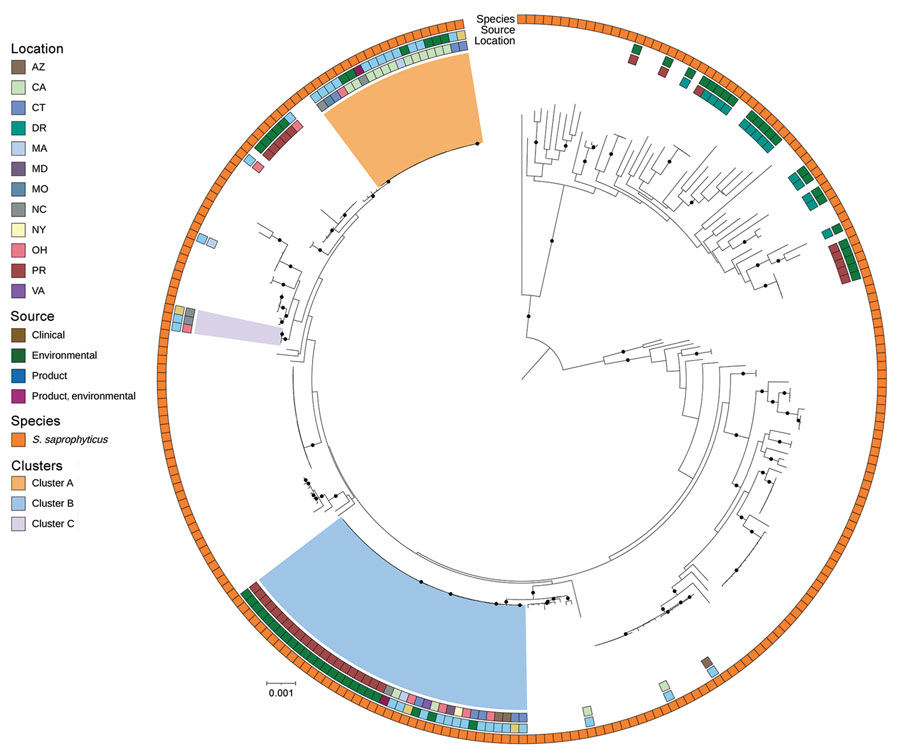
Figure 5. Public Staphylococcus saprophyticus genomes and study isolates from investigation of bacterial contamination of platelet blood products, United States, 2018–2022. Shown is a RaxML (https://cme.h-its.org)‒generated phylogeny based on 1,808 core genes of all S. saprophyticus isolates from this study and all S.saprophyticus genomes from the National Center for Biotechnology Information RefSeq (https://www.ncbi.nlm.nih.gov/refseq) database. Isolate location, isolate source, and species from National Center for Biotechnology Information or by average nucleotide identity were layered onto the phylogeny. Light orange, blue, and purple indicate the 3 clusters from Figure 3, panel A. Black circles on branches indicate 100% support for the branch of 100 bootstraps. US states are identified by 2-letter postal codes. Scale bar indicates nucleotide substitutions per site. DR, Dominican Republic; PR, Puerto Rico.
Main Article
Page created: August 02, 2023
Page updated: September 20, 2023
Page reviewed: September 20, 2023
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
