Evolution of Avian Influenza Virus (H3) with Spillover into Humans, China
Jiaying Yang
1, Ye Zhang
1, Lei Yang
1, Xiyan Li, Hong Bo, Jia Liu, Min Tan, Wenfei Zhu, Yuelong Shu

, and Dayan Wang

Author affiliations: National Institute for Viral Disease Control and Prevention, Chinese Center for Disease Control and Prevention, Key Laboratory for Medical Virology, Beijing, China (J. Yang, Y. Zhang, L. Yang, X. Li, H. Bo, J. Liu, M. Tan, W. Zhu, D. Wang); School of Public Health (Shenzhen), Shenzhen campus of Sun Yat-sen University, Shenzhen, China (J. Yang, Y. Shu); Institute of Pathogen Biology of Chinese Academy of Medical Science/Peking Union Medical College, Beijing (Y. Shu)
Main Article
Figure 3
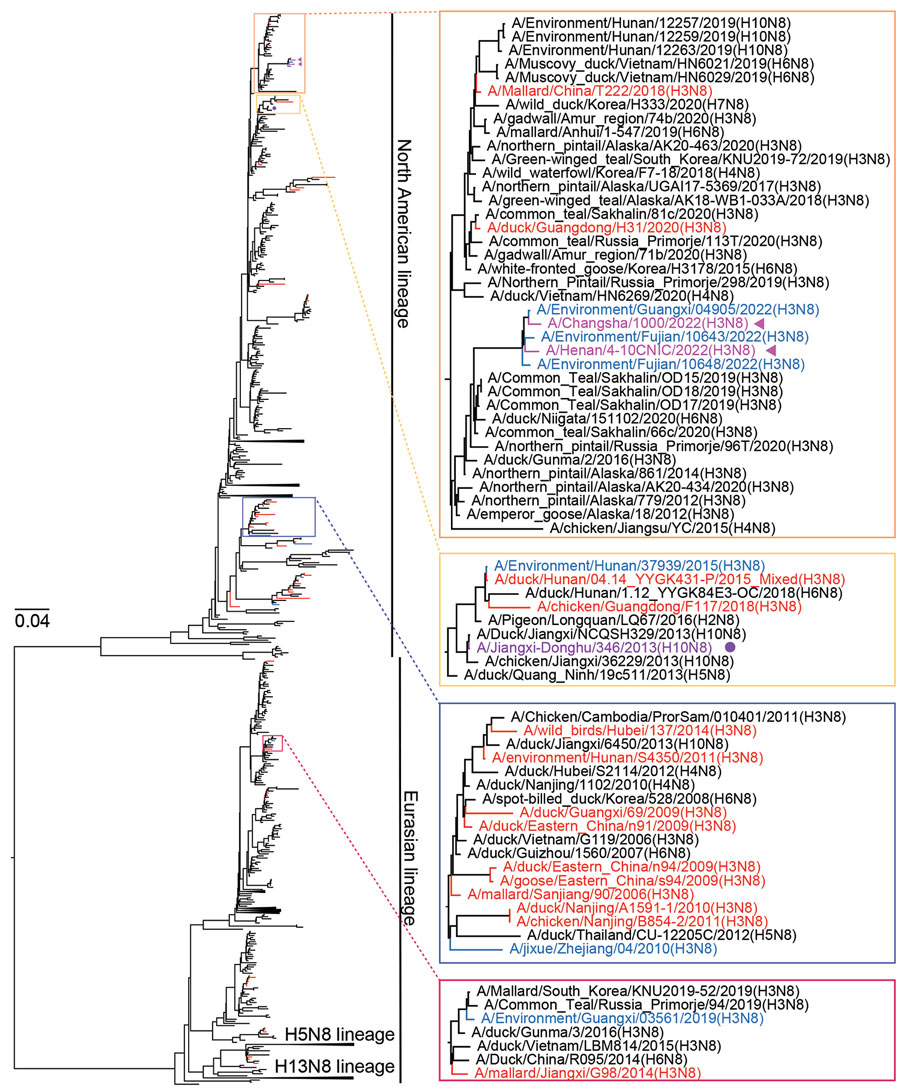
Figure 3. Maximum-likelihood phylogenetic tree of avian influenza virus subtype N8 genes from China (n = 1,106) and reference sequences from GISAID (https://www.gisaid.org). Blue tree sections indicate sequences of H3 subtype viruses reported in this study; red tree sections indicate other H3 subtype viruses from China. For of clarity, some clades are collapsed. Representative clusters are indicated in shaded boxes and magnified on the right. Violet arrows indicate human H3N8 viruses; purple solid circle indicates human H10N8 virus. The phylogenetic tree of N8 genes with more complete information is shown in Appendix Figure 2, panel D). Scale bar indicates nucleotide substitutions per site.
Main Article
Page created: April 06, 2023
Page updated: May 17, 2023
Page reviewed: May 17, 2023
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
