Volume 29, Number 6—June 2023
Research
Evolution of Avian Influenza Virus (H3) with Spillover into Humans, China
Figure 5
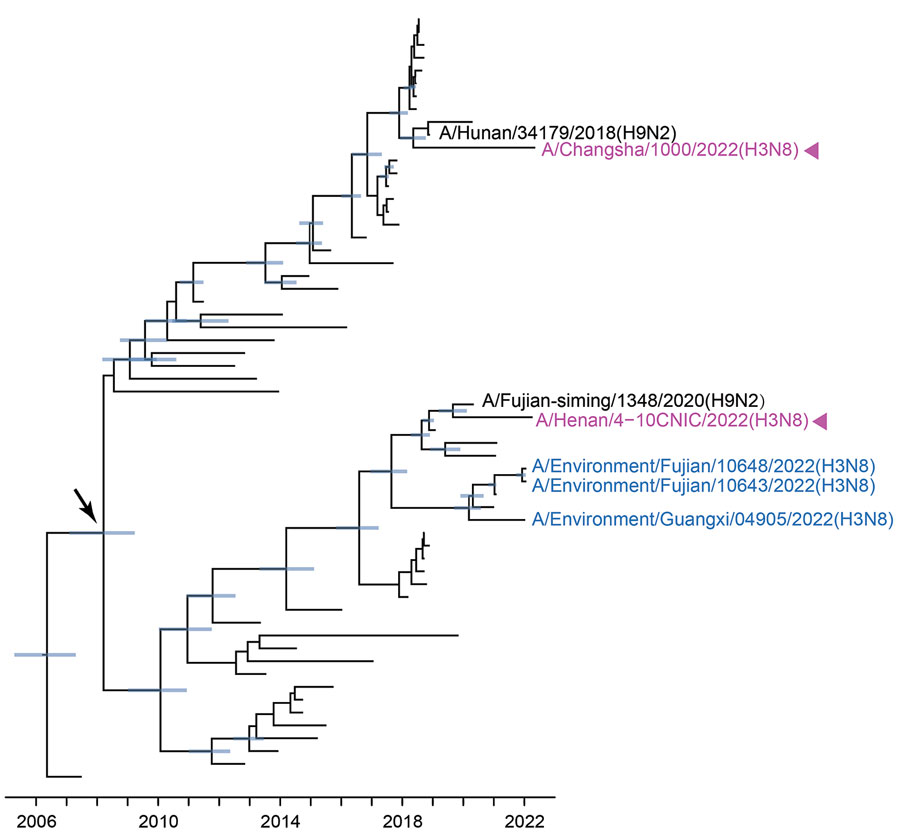
Figure 5. Bayesian time-resolved phylogenetic tree of polymerase basic 1 genes (n = 60) from avian influenza viruses subtype H3 from China and reference sequences from GISAID (https://www.gisaid.org). Violet indicates human H3N8 viruses; blue indicates H3N8 G25 AIVs sequenced in this study; black indicates the closest strains to human H3N8 viruses. Blue node bars correspond to the 95% credible intervals of node heights. Arrow indicates the most recent common ancestor of H3N8 G25 viruses. The fully resolved tree with detailed information is depicted in Appendix Figure 9.
1These authors contributed equally to this article.
Page created: April 06, 2023
Page updated: May 17, 2023
Page reviewed: May 17, 2023
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.