Volume 30, Number 12—December 2024
Research
Novel Mastadenovirus Infection as Cause of Pneumonia in Imported Black-and-White Colobuses (Colobus guereza), Thailand
Cite This Article
Citation for Media
Abstract
We identified a novel mastadenovirus, herein referred to as colobus adenovirus (CoAdV), as the likely cause of fatal respiratory and enteric diseases in multiple black-and-white colobuses (Colobus guereza) imported into Thailand in 2022. Among 9 affected colobuses, 4 died. Viral antigen was abundant in respiratory and enteric tissues, where prominent lesions and clinical signs were observed. We successfully cultivated CoAdV in Vero cells and characterized the complete viral genome, which indicated the virus is genetically distinct from other simian adenoviruses. We also conducted a retrospective study of archival samples from 7 other unrelated colobuses that had respiratory distress or diarrhea and found similar viral strains in 4 of those colobuses. Although we could not determine the potential harm to humans or other nonhuman primates from current information, the zoonotic and spillover potential of this virus to other related hosts should not be neglected. Veterinarians should consider CoAdV when pneumonia is diagnosed in colobuses.
Most adenoviruses that infect mammals belong to the genus Mastadenovirus, which is part of the family Adenoviridae and includes a variety of viruses found in humans, nonhuman primates (NHPs), and other mammals (1,2). Although many cases of human adenovirus infection are predominantly asymptomatic and infection can be self-limited in immunocompetent hosts, infections in immunosuppressed patients can lead to greater severity and even death (3–6).
Of the >50 adenovirus species in the genus Mastadenovirus, 9 that infect NHPs, known as simian adenoviruses (SAdVs; SAdV-A–I), and 7 that infect humans, known as human adenoviruses (HAdVs; HAdV-A–G), have been documented (1). Although species domination is considered on the basis of infected hosts, many adenoviruses found in NHPs have been identified as HAdVs (7), and SAdV infections have also been reported in humans who have contact with NHPs (6). Because of the phylogenetic proximity between NHPs and humans, interspecies transmission is highly possible (8,9). Although adenoviruses have been considered host-specific and restricted to >1 related species (4), phylogenetic and evolutionary analyses reveal instances of interspecies transmission that have led to new host-virus adaptations. For example, HAdV-E is believed to have originated from chimpanzee adenoviruses and HAdV-B from gorilla adenoviruses (10). In addition, serologic evidence indicates that adenoviruses derived from monkeys can infect humans who have close contact with NHPs (5,6). Those findings raise concerns about interspecies transmission and zoonotic potential. Notable examples include canine adenovirus 1, which infects multiple carnivore species, and skunk adenovirus 1, found in marmosets (Callithrix spp.), skunks (Mephitis spp.), pygmy hedgehogs (Atelerix albiventris), and other species, illustrating adenoviruses’ ability to cross species barriers (11,12). Such instances underscore the potential zoonotic threats posed by mastadenoviruses because of their genetic diversity and adaptability (13).
Southeast Asia and China are main hubs of the primate trade, both as importers and exporters, for various purposes (14,15). Importation of apparently healthy NHPs without specific surveillance for NHP-related adenoviruses may result in the introduction of novel adenoviruses to naive environments or vice versa. We identified a novel adenovirus in colobuses with fatal respiratory distress that were imported into Thailand in 2022.
During August–September 2022, nine wild-caught adult black-and-white colobuses (Colobus guereza) of unknown specific ages were imported from South Africa and kept as pets in individual stainless-steel cages in Bangkok, Thailand. All animals were shipped together in a single lot and underwent quarantine at customs for several months before all 9 went to a single pet owner. Initially, clinical signs and symptoms began with depression, reduced appetite, and fever, after which coughing, pneumonia, and subsequent respiratory distress developed. Those symptoms were observed shortly (within a few weeks) after the animals arrived at the final destinations with the pet owner. Four of the colobuses (nos. 1–4) succumbed to their illnesses 5–10 days after the onset of clinical symptoms; the other 5 (nos. 5–9) survived after receiving intensive supportive care.
Clinical samples, including oral and rectal swab samples, were collected from all 9 diseased colobuses. Among the 9 colobuses, the 4 that subsequently died (nos. 1–4) were submitted for necropsy at the Department of Pathology, Faculty of Veterinary Science, Chulalongkorn University, Bangkok. We collected vital samples, including brain, trachea, lung, heart, intestine, liver, kidneys, and lymph nodes, and divided samples into 2 cohorts: fresh-frozen tissues and formalin-fixed paraffin-embedded (FFPE) tissues fixed in 10% buffered formalin. This study was approved by Chulalongkorn University Animal Care and Use Committee (approval no. 2431048).
Nucleic Acid Extraction and Routine Diagnostic Panels
We processed collected samples for viral nucleic acid extraction by using QIAamp Viral RNA Mini Kit (QIAGEN, https://www.qiagen.com) according to the manufacturer’s guidelines. To elucidate the viral infections in these cases, we subjected the extracted nucleic acid samples to routine virologic testing panels by using pan-family virologic PCRs targeting Herpesviridae (16), Paramyxoviridae (17), Pneumoviridae (17), Parvoviridae (18), Orthomyxoviridae (19), Retroviridae (20), and Adenoviridae (21) families. For PCR-positive samples, we sent target amplicons to Celemics, Inc. (https://www.celemics.com) for next-generation sequencing (NGS)–based methods. Furthermore, we submitted fresh lung samples from all investigated colobuses for aerobic bacterial culture.
Detection of Adenovirus DNA and Complete Genome Characterization
For samples that initially tested positive for adenoviruses by pan-Adenoviridae PCR targeting the DNA polymerase (DNApol) gene, we reconfirmed results by using a second PCR that targeted the hexon gene with established pan-Mastadenovirus primers (22). We then sequenced the positive amplicons following the protocol described previously. We used nucleic acid extracts obtained from fresh lung tissue of colobus no. 1 for complete adenoviral genome characterization through a de novo–based NGS method (Vishuo Biomedical, https://www.vishuo.com) (Appendix). Subsequently, we aligned the identified sequence with previously described adenovirus sequences found in NHPs, including SAdV-A–I and unclassified SAdVs. We subjected that alignment to maximum-likelihood phylogenetic analysis (Appendix). We also performed recombination analysis by using a recombination detection program (23) (Appendix). We refer to this novel virus as colobus adenovirus (CoAdV).
Viral Load Quantification and Localization
To confirm the results from NGS analysis and investigate the genomic distribution of CoAdV in various organs and in other colobuses, we conducted conventional PCR using newly designed primers targeting the DNApol gene of CoAdV. In addition, to quantify viral loads in different organs, we performed a SYBR green–based quantitative PCR (qPCR) by using KAPA SYBR Fast qPCR Master Mix (2X) Universal Kit (Sigma-Aldrich, https://www.sigmaaldrich.com) with specific primers designed to target the DNApol gene of CoAdV (CoAdV-DpolF-6348, 5′-CAG CTG GTC CTC GTC C-3′; CoAdV-DpolR-6443, 5′-TTG CAG GAC CCC CTG AAG AC-3′). We estimated relative viral loads in each organ on the basis of cycle threshold (Ct) values.
Detection of CoAdV Antigen in FFPE Tissues
To determine the tissue tropism of CoAdV, we subjected FFPE tissues from the 4 necropsied colobuses that tested positive in both conventional and qPCR to immunohistochemical (IHC) analysis. The examined tissues were brain, lung, liver, lymph node, spleen, heart, kidneys, and intestine. We assessed localization of CoAdV in those tissues by using a horseradish peroxidase polymer-conjugated method with primary antibody against adenovirus clones 2/6 and 20/11 (Chemi-Con, https://chemi-con.com) (Appendix).
Viral Isolation
We conducted virus isolation on Vero cells (ATCC accession no. CCL-81) in 12-well tissue culture plates. We subjected samples to viral isolation, including oral and rectal swab samples from diseased colobus nos. 4–6, 7, and 9 that had been collected in viral transport medium, and supernatants from lung and trachea homogenized in PBS from colobus no. 1 that died (Appendix). We monitored cultures for cytopathic effects (CPE) daily for 5 days (2). We collected supernatants from wells showing CPE and stored at –80°C for subsequent adenovirus-specific PCR confirmation. In addition, we harvested cell pellets and prepared for transmission electron microscopy analysis.
Retrospective Study of CoAdV in Other NHPs
We also used the qPCR described to conduct CoAdV detection on a small sample cohort that underwent postmortem examination at Department of Pathology, Faculty of Veterinary Science, Chulalongkorn University, during 2020–2022. The cohort comprised archival respiratory or rectal swab samples and fresh-frozen lung tissues obtained from 7 other colobuses that died from various causes, plus the fresh-frozen lung or respiratory swab samples from other primates, including 2 Douc monkeys (Pygathrix spp.), 1 De Brazza’s monkey (Cercopithecus neglectus), 6 marmosets, 4 Japanese macaques (Macaca fuscata), 2 cotton-top tamarins (Saguinus oedipus), and 11 long-tailed macaques (Macaca fascicularis).
CoAdV Detection, Genome Characterization, and Phylogenetic Analysis
Apart from negative results of ancillary testing using pan-family virologic PCR panels, the oral swab samples from all 9 colobuses and 5 fecal swab samples obtained from colobuses nos. 1–4 and 7 tested positive by pan-adenovirus PCR. The colobuses that subsequently died (nos. 1–4) predominantly exhibited lower Ct values (Table). Analysis of tissue samples showed that the lung, trachea, and intestines exhibited positive amplification signals for the pan-adenovirus PCR, whereas the other collected tissues had negative results. Klebsiella spp. was cultivated from the lung tissue of colobus nos. 1 and 4. We sequenced the positive adenovirus amplicons (460 bp) from all samples, and initial BLASTn (https://blast.ncbi.nlm.nih.gov) results showed 99.9% nucleotide similarities among adenovirus from colobuses and most were closely related to another SAdV (GenBank accession no. JN808448).
NGS results showed a total of 889 sequence reads that mapped to various simian mastadenovirus species. From those sequence reads, we generated a consensus sequence of 33,813 bp, tentatively named CoAdV strain CP001-TH/2023. To confirm the accuracy of the generated CoAdV sequence, we used primers for adenovirus screening as described and submitted the 460-bp PCR products for Sanger sequencing, which showed 100% similarity to the generated sequence. The CoAdV genome comprises 19.85% A, 18.34% T, 30.80% C, and 31.01% G. The CoAdV strain CP001-TH/2023 genome contained 145-bp ends of inverted terminal repeats with a conserved motif CATCATCCAAT, displaying typical mastadenovirus orthologs. The CoAdV CP001-TH/2023 genome possessed 32 predicted proteins and included genus-specific genes encoding proteins V and IX and the early (E) regions E1, E3, and E4. Only one fiber gene was observed within the CoAdV CP001-TH/2023 genome (Figure 1). We submitted the CoAdV sequence to GenBank (accession no. PP985428).
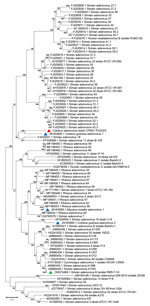
The full-length CoAdV strain CP001-TH/2023 exhibited 67.10% nucleotide similarity to the SAdV-6 strain SV-39 (GenBank accession no. JQ776547) from Old World macaque monkeys in the United States. CoAdV CP001-TH/2023 was distinct from other SAdVs, and nucleotide similarities ranged from 66.50% to 50.02%. The human mastadenovirus B isolate KUMC-62 (GenBank accession no. KY320276) was the most distant strain, showing only 50.02% nucleotide similarity to the detected CoAdV. Analysis of DNApol gene revealed nucleotide similarities of 62.30%–77.90% and amino acid similarities of 25.40%–57.6% to previously described SAdVs. The SAdV strain ER (GenBank accession no. MZ062897) identified in China was the most genetically similar strain. Phylogenetic analysis of the full-length genome of SAdVs revealed that CoAdV CP001-TH/2023 formed a distinct phylogenetic cluster, and was most closely related to SAdV-A, SAdV-G, and SAdV-I isolated from Old World monkeys (Figure 2) and that SAdV-6 strain SV-39 was the most closely related. The whole-genome phylogenetic tree was consistent with the phylogenetic analysis of amino acid sequences of the DNApol and IVa2 trees (Figures 3, 4).
Because limited information was available on other genes of previously described AdVs identified in colobuses in Germany, we were only able to analyze relatedness in the fiber gene (Figure 5). CoAdV CP001-TH/2023 shared 71.75%–80.50% nucleotide similarity with AdVs identified in colobuses in Germany in 2011, and Colobus guereza adenovirus 1 (GenBank accession no. JN163994) was the most closely related strain. Although the CoAdV hexon gene identified in this study genetically clustered closely with the Colobus guereza adenovirus 1 (GenBank accession no. JN163994) strain, it formed a distant phylogenic topology with 99% bootstrap support (Figure 6). However, CoAdV CP001-TH/2023 had a unique phylogenetic topology and did not cluster with other previously described AdVs identified in colobuses (GenBank accession nos. JN613993 and JN613995). Information for complete genome sequences of SAdV identified in Thailand was limited, and only the fiber gene was available. Thus, we also included the fiber gene of SAdV-B isolate RBR-7-10 (GenBank accession no. ON072488) to determine the phylogenetic relationship of the detected CoAdV; however, those sequences were distantly related (Figure 5). We did not identify any genetic recombination events within the CoAdV genome.
CoAdV Pathology and Viral Distribution
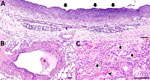
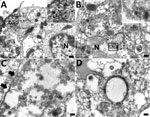
Overall, histologic findings were consistent across all colobuses and showed varying degrees of severity. The trachea mucosa was extensively replaced by laminated bands of eosinophilic fibrillar material mixed with scant aggregates of karyorrhectic debris. In addition, the tracheal lumen contained a small pool of neutrophils and karyorrhectic debris (Figure 7, panel A). The pulmonary interstitium was diffusely congested, with increased prominence and tortuosity of markedly engorged alveolar capillaries. In colobus no. 1, the pulmonary alveoli were multifocally filled with eosinophilic granular to homogenous material, intermixed with variable numbers of foamy macrophages, and fewer polymorphonuclear cells (Figure 7, panel B). Moreover, alveoli were overlaid by variable flocculent mats of eosinophilic fibrin. Similar mats of eosinophilic fibrin and a few neutrophils were seen in 1 lumen of the large bronchial airways (Figure 7, panel C). In colobus no. 2, we noted similar but milder changes.
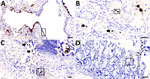
To determine the CoAdV distribution in tissue and localize the virus where lesions were located, we quantified the viral loads by using qPCR (Table). We primarily detected CoAdV in respiratory samples and noted the highest viral loads in the lung but also high loads in trachea tissue and oral and fecal swab samples. We found smaller amounts of the viral genome in the small intestine. We used IHC to determine CoAdV tissue localization, which revealed prominent positive immunoreactivity in the nuclei of bronchial epithelial cells (Figure 8, panel A), pulmonary alveolar macrophages, and endothelial cells of pulmonary capillaries (Figure 8, panel B). We also found local immunolabelling in the mucosal epithelial cells and glands of the trachea, particularly in areas that had pseudodiphtheritic membrane of necrosis (Figure 8, panel C). We identified IHC labeling in the small intestine (Figure 8, panel D) and in single lymphoid cells in mediastinal lymph node and tonsil.
Viral Culture
We successfully isolated CoAdV from fecal and oral swab samples of colobus nos. 6, 7, and 9 and from homogenized trachea and lung tissues of colobus no. 1. However, cell cultures inoculated with samples from colobuses 4 and 5 exhibited severe cytotoxicity, characterized by cell detachment and cell death at the first day postinoculation (dpi), leading us to discard those inoculations. Infected Vero cells revealed nonsyncytia, round-shaped CPE (Figure 9). The CPE first appeared at 3 dpi, characterized by cell rounding and detachment observed by 5 dpi. Subsequent infection of Vero cells with the supernatant from the primary virus isolation also resulted in similar CPE formation at 3 dpi. Conventional PCR confirmed the presence of CoAdV DNA in the infected cell culture supernatants. Sanger sequencing of the partial DNApol gene from cellular supernatant identified partial CoAdV sequences. We did not detect CoAdV DNA in mock-infected cells.
In Situ Ultrastructure of CoAdV
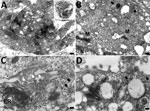
Ultrastructural investigation of CoAdV-infected FFPE sections revealed pulmonary cellular damage by exhibiting rupture of cellular membrane, cytoplasmic vacuolization, and degenerated nuclear compartments. Within intact cells, condensed nuclear membrane contained various sizes of large cytoplasmic vacuolization containing numerous icosahedral electron-dense particles, each ≈80 nm in diameter, characteristic of adenoviruses (Figure 10, panel A). We considered those particles, typically seen attached near ruptured nuclear membranes or distributed randomly within the cytosol (Figure 10, panels B–D), of viral origin. Transmission electron microscopy analysis of CoAdV-infected Vero cells at 1 dpi showed prominent nuclear chromatin with early detection of viral virion within the nucleus (Figure 11, panel A). We also noted vesicles containing 80–90 nm icosahedral electron-dense or electron-lucent particles. We found cytoplasmic vacuolization and disruption of nuclear membranes in infected Vero cells at 5 dpi. We frequently observed numerous electron-dense particles in the cytosol. Virions were scattered throughout the cytosol and found within the endoplasmic reticulum or Golgi apparatus (Figure 11, panel B). In some infected cells, we observed mature virions fused with the cellular membrane being released into the extracellular space or passively through ruptured plasma membranes (Figure 11, panels C, D).
Retrospective Study
We detected CoAdV nucleic acid in 4 oral and 2 rectal swab samples collected and archived from 4 of 7 colobuses that were imported into Thailand, exhibited respiratory distress, and were submitted for postmortem investigation during 2020–2022. Phylogenetic analysis of obtained sequences revealed that the sequences clustered together and were closely related to the CoAdV identified in this study (Appendix Figure). Investigation of archival samples derived from various other simian species were negative.
Using conventional PCR targeting the conserved DNApol gene of adenoviruses, we identified a novel mastadenovirus, CoAdV, in multiple black-and-white colobuses, a species currently listed as decreasing in population (24). Identification of this virus was associated with a fatal outbreak of respiratory disease. We propagated CoAdV in Vero cells and confirmed the virus via PCR and ultrastructural investigation. qPCR quantified viral loads in various organs, and immunohistochemistry and transmission electron microscopy revealed virus localization in tracheal epithelium and pulmonary parenchyma, associated with pneumonia.
We successfully isolated CoAdV and characterized its complete genome sequence. Subsequent tissue analysis revealed that CoAdV infection was linked to respiratory lesions. Detection of CoAdV DNA and its localization in tissues with lesions suggests a major disease burden. Moreover, the identification of CoAdV in colobuses within the same household may indicate the virus’ contagious nature. Therefore, CoAdV infection should be considered a concern when pneumonia occurs in this species.
Adenovirus infection in colobuses has been reported previously (9), but no information regarding clinical relevance was provided in those cases, and only the partial hexon gene was characterized. Although phylogenetic analysis of the hexon gene revealed that our CoAdV CP001-TH/2023 is genetically close to Colobus guereza adenovirus 1 (GenBank accession no. JN163994), the virus we detected still exhibits diversity and shares less genomic and amino acid similarity with that virus.
Infection associated with fatal outcomes in these colobuses cannot be definitively concluded without conducting animal challenge studies. The clinical severity of adenovirus infection in NHPs varies and often is associated with fatal diseases when infecting immunocompromised subjects (4). Although metagenomic studies in the fatal CoAdV cases we report did not indicate other underlying viral infection and results of ancillary testing were negative, the severe clinical outcomes observed suggest a potential role for stress-induced reactivation of a latent virus in the colobuses, exacerbated by transportation stress and possibly inadequate quarantine measures. In addition, the widespread severity of the illnesses raises the possibility that CoAdV might have originated from another animal source and was not native to this species. Although our phylogenetic analysis did not confirm direct links to viruses in related hosts, the adaptability of adenoviruses across species barriers supports that hypothesis (6,7,10,11).
To determine whether CoAdV is circulating or originating in Thailand, we made further attempts to investigate CoAdV infection in other NHPs; however, we could not identify this virus in samples from other NHPs, except in other colobuses that had respiratory problems or diarrhea. Further studies are needed to assess whether CoAdV is host-restricted because the infection appears potentially limited to colobuses. Detection of CoAdV in archival samples suggests the possibility of longstanding circulation within the colobus population. Supporting previous publications (2,25), the adenovirus identified in the colobuses was not genetically related to previously described adenoviruses in free-ranging macaques in Thailand. However, because we only investigated a small number of samples, definitive conclusions cannot be drawn. Future investigations incorporating serologic surveys could provide a more comprehensive assessment of the virus’ prevalence and transmission patterns. Such studies would be particularly valuable for determining whether CoAdV is circulating endemically or was recently introduced into Thailand. Because samples from close contact humans have not been investigated, the potential harmful effects of CoAdV infection to humans cannot be extrapolated. Reports of SAdV infection in non–natural hosts and humans, and subsequent transmission to other contracts (6), highlight the zoonotic transmission risk. Therefore, the spillover of CoAdV to susceptible hosts, including humans, should not be overlooked.
The CoAdV genome contains 32 putative coding sequences, including unique genes such as V and IX, and the E1, E3, and E4 regions that are specific to the Mastadenovirus genus (1). CoAdV also has a single fiber gene, which is uncommon in related viruses. The <85% amino acid identity to its closest relative, SAdV-6 strain SV-39, and its identification in a new host support classifying CoAdV as a distinct species. Thus, we propose the virus as SAdV-J on the basis of species demarcation.
CoAdV phylogenetically clustered with most mastadenoviruses found in old-world monkeys, confirming unique clusters of adenoviruses within the Cercopithecidae family. Phylogenetic analysis of DNApol gene revealed that CoAdV shares genetic similarity with SAdV strain 55, identified in an endangered golden sub-nosed monkey (Rhinopithecus roxellana) in China, suggesting that CoAdV may have co-evolved with that viral SAdV species. Although the partial hexon gene of adenoviruses identified in colobuses in Germany was reported (9), our phylogenetic analysis of the hexon gene indicated that CoAdV is not related to those former strains. Given the unique phylogenic topology of CoAdV and the fact that interspecies recombination of mastadenoviruses has been previously reported (8,9), we investigated the potential recombination events. However, we did not identify any recombination breakpoints within the CoAdV genome. Although we successfully characterized a complete genome sequence of CoAdV, we acknowledge that the genomic analysis was limited to that single sequence. Future studies should aim to include multiple genome sequences to enhance our understanding of the genetic diversity and epidemiology of CoAdV.
Colobuses, native to Africa, are now experiencing a population decline (24). Despite extensive efforts to protect this species from becoming endangered, CoAdV infection causing fatal disease poses a possible threat. The transmission of CoAdV to native monkeys has not been elucidated, and the natural host of this virus remains unidentified, although imported infected reservoirs may spread the infection.
In conclusion, our study demonstrates infection with a novel virus, CoAdV, associated with respiratory disease in imported black-and-white colobuses and describes the isolation and characterization of this virus. CoAdV was identified in all colobus contacts, indicating its transmissibility. Genetic analysis of the isolated virus revealed that CoAdV is distantly related to other previously described SAdVs and appears to be a novel species in the genus Mastadenovirus. Although we did not find CoAdV in retrospective samples obtained from other NHPs, the potential for infection in NHPs and humans remains unclear. Further studies are needed to determine the virus’ role in disease development and host diversity. Extensive AdV surveillance in various animal species is crucial for elucidating the origins and spread of CoAdV in colobuses.
Dr. Piewbang is a senior postdoctoral fellow at the Department of Pathology, Faculty of Veterinary Science, Chulalongkorn University, Bangkok, Thailand. His research interests include virus discovery, emerging and re-emerging infections, cross-species transmission, and viral evolution.
Acknowledgments
We thank Patcharee Umroong for excellent transmission electron microscopy sample preparation.
This research project was supported by Ratchadapisek Somphot Fund for Postdoctoral Fellowship, Chulalongkorn University to C.P.; National Research Council of Thailand grant no. NRCT5-RGJ63001-013 and The Second Century Fund (C2F), Chulalongkorn University to P.P.; National Research Council of Thailand (grant no. N41A640189) and The Second Century Fund (C2F), Chulalongkorn University to P.L.; The Second Century Fund (C2F), Chulalongkorn University to S.W.W.; National Research Council of Thailand R. Thanawongnuwech TRF Senior Scholar 2022 grant no. N42A650553 and Animal Virome and Diagnostic Development Research Unit, Faculty of Veterinary Science, Chulalongkorn University, to S.T.
References
- Benkő M, Aoki K, Arnberg N, Davison AJ, Echavarría M, Hess M, et al.; Ictv Report Consortium. ICTV virus taxonomy profile: Adenoviridae 2022. J Gen Virol. 2022;103:
001721 . DOIPubMedGoogle Scholar - Kosoltanapiwat N, Tongshoob J, Ampawong S, Reamtong O, Prasittichai L, Yindee M, et al. Simian adenoviruses: Molecular and serological survey in monkeys and humans in Thailand. One Health. 2022;15:
100434 . DOIPubMedGoogle Scholar - Wachtman LM, Mansfield KG. Opportunistic infections in immunologically compromised nonhuman primates. ILAR J. 2008;49:191–208. DOIPubMedGoogle Scholar
- Rogers DL, Ruiz JC, Baze WB, McClure GB, Smith C, Urbanowski R, et al. Epidemiological and molecular characterization of a novel adenovirus of squirrel monkeys after fatal infection during immunosuppression. Microb Genom. 2020;6:
mgen000395 . DOIPubMedGoogle Scholar - Chiu CY, Yagi S, Lu X, Yu G, Chen EC, Liu M, et al. A novel adenovirus species associated with an acute respiratory outbreak in a baboon colony and evidence of coincident human infection. MBio. 2013;4:e00084–13. DOIPubMedGoogle Scholar
- Chen EC, Yagi S, Kelly KR, Mendoza SP, Tarara RP, Canfield DR, et al. Cross-species transmission of a novel adenovirus associated with a fulminant pneumonia outbreak in a new world monkey colony. PLoS Pathog. 2011;7:
e1002155 . DOIPubMedGoogle Scholar - Medkour H, Amona I, Akiana J, Davoust B, Bitam I, Levasseur A, et al. Adenovirus infections in African humans and wild non-human primates: great diversity and cross-species transmission. Viruses. 2020;12:657. DOIPubMedGoogle Scholar
- Tan B, Wu LJ, Yang XL, Li B, Zhang W, Lei YS, et al. Isolation and characterization of adenoviruses infecting endangered golden snub-nosed monkeys (Rhinopithecus roxellana). Virol J. 2016;13:190. DOIPubMedGoogle Scholar
- Wevers D, Metzger S, Babweteera F, Bieberbach M, Boesch C, Cameron K, et al. Novel adenoviruses in wild primates: a high level of genetic diversity and evidence of zoonotic transmissions. J Virol. 2011;85:10774–84. DOIPubMedGoogle Scholar
- Hoppe E, Pauly M, Gillespie TR, Akoua-Koffi C, Hohmann G, Fruth B, et al. Multiple cross-species transmission events of human adenoviruses (HAdV) during hominine evolution. Mol Biol Evol. 2015;32:2072–84. DOIPubMedGoogle Scholar
- Needle DB, Selig MK, Jackson KA, Delwart E, Tighe E, Leib SL, et al. Fatal bronchopneumonia caused by skunk adenovirus 1 in an African pygmy hedgehog. J Vet Diagn Invest. 2019;31:103–6. DOIPubMedGoogle Scholar
- Balboni A, Verin R, Morandi F, Poli A, Prosperi S, Battilani M. Molecular epidemiology of canine adenovirus type 1 and type 2 in free-ranging red foxes (Vulpes vulpes) in Italy. Vet Microbiol. 2013;162:551–7. DOIPubMedGoogle Scholar
- Borkenhagen LK, Fieldhouse JK, Seto D, Gray GC. Are adenoviruses zoonotic? A systematic review of the evidence. Emerg Microbes Infect. 2019;8:1679–87. DOIPubMedGoogle Scholar
- Ni Q, Wang Y, Weldon A, Xie M, Xu H, Yao Y, et al. Conservation implications of primate trade in China over 18 years based on web news reports of confiscations. PeerJ. 2018;6:
e6069 . DOIPubMedGoogle Scholar - Nekaris KAI, Bergin D. Primate trade (Asia). In: Fuentes A, editor. The international encyclopedia of primatology. Hoboken (NJ, USA): John Wiley & Sons; 2017. p. 1–8.
- VanDevanter DR, Warrener P, Bennett L, Schultz ER, Coulter S, Garber RL, et al. Detection and analysis of diverse herpesviral species by consensus primer PCR. J Clin Microbiol. 1996;34:1666–71. DOIPubMedGoogle Scholar
- Tong S, Chern SW, Li Y, Pallansch MA, Anderson LJ. Sensitive and broadly reactive reverse transcription-PCR assays to detect novel paramyxoviruses. J Clin Microbiol. 2008;46:2652–8. DOIPubMedGoogle Scholar
- Mochizuki M, Horiuchi M, Hiragi H, San Gabriel MC, Yasuda N, Uno T. Isolation of canine parvovirus from a cat manifesting clinical signs of feline panleukopenia. J Clin Microbiol. 1996;34:2101–5. DOIPubMedGoogle Scholar
- Tong S, Li Y, Rivailler P, Conrardy C, Castillo DA, Chen LM, et al. A distinct lineage of influenza A virus from bats. Proc Natl Acad Sci U S A. 2012;109:4269–74. DOIPubMedGoogle Scholar
- Hwa CZR, Tsai SP, Yee JL, Van Rompay KK, Roberts JA. Evidence of simian retrovirus type D by polymerase chain reaction. J Med Primatol. 2017;46:79–86. DOIPubMedGoogle Scholar
- Wellehan JF, Johnson AJ, Harrach B, Benkö M, Pessier AP, Johnson CM, et al. Detection and analysis of six lizard adenoviruses by consensus primer PCR provides further evidence of a reptilian origin for the atadenoviruses. J Virol. 2004;78:13366–9. DOIPubMedGoogle Scholar
- Crawford-Miksza LK, Nang RN, Schnurr DP. Strain variation in adenovirus serotypes 4 and 7a causing acute respiratory disease. J Clin Microbiol. 1999;37:1107–12. DOIPubMedGoogle Scholar
- Poonsin P, Wiwatvisawakorn V, Chansaenroj J, Poovorawan Y, Piewbang C, Techangamsuwan S. Canine respiratory coronavirus in Thailand undergoes mutation and evidences a potential putative parent for genetic recombination. Microbiol Spectr. 2023;11:
e0226823 . DOIPubMedGoogle Scholar - de Jong YA, Butynski TM, Oates JF. Colobus guereza. In: International Union for Conservation of Nature and Natural Resources, editor. The IUCN Red List of threatened species 2019 [cited 2024 Jun 16].
- Kosoltanapiwat N, van der Hoek L, Kinsella CM, Tongshoob J, Prasittichai L, Klein M, et al. A novel simian adenovirus associating with human adeno-virus species G isolated from long-tailed macaque feces. Viruses. 2023;15:1371. DOIPubMedGoogle Scholar
Figures
Table
Cite This ArticleOriginal Publication Date: November 21, 2024
Table of Contents – Volume 30, Number 12—December 2024
| EID Search Options |
|---|
|
|
|
|
|
|





Please use the form below to submit correspondence to the authors or contact them at the following address:
Somporn Techangamsuwan, Department of Pathology, Faculty of Veterinary Science, Chulalongkorn University, Pathumwan, Bangkok 10330, Thailand
Top