Volume 30, Supplement—October 2024
SUPPLEMENT ISSUE
Articles
Etiology and Epidemiology of Travelers’ Diarrhea among US Military and Adult Travelers, 2018–2023
Abstract
Travelers’ diarrhea has a high incidence rate among deployed US military personnel and can hinder operational readiness. The Global Travelers’ Diarrhea study is a US Department of Defense–funded multisite surveillance effort to investigate the etiology and epidemiology of travelers’ diarrhea. During 2018–2023, we enrolled 512 participants at partner institutions in 6 countries: Djibouti, Georgia, Egypt, Honduras, Nepal, and Peru. Harmonized laboratory methods conducted at each partner institution identified >1 pathogens, including Escherichia coli (67%–82%), norovirus (4%–29%), and Campylobacter jejuni (2%–20%), in 403 (79%) cases. Among cases, 79.7% were single infections, 19.6% were double infections, and 0.7% were triple infections. The most common enterotoxigenic E. coli colonization factors identified were CS3 (25%) and CS21 (25%), followed by CS2 (18%) and CS6 (15%). These data can inform best treatment practices for travelers’ diarrhea and support US military health readiness.
Travelers’ diarrhea (TD) is a gastrointestinal (GI) illness that affects millions of people each year, and infection rates range from 30% to 70% among travelers within 2 weeks of travel initiation, depending upon geographic region and seasonality of travel (1,2). Symptoms can range from mild cramps and loose stool to bloody diarrhea, fever, abdominal pain, and vomiting. Bacterial pathogens are the leading causative agents of TD, accounting for >80% of cases (1).
In March 2023, the United States Military Infectious Disease Research Panel’s Threat Prioritization Panel determined that bacterial diarrhea was the number 1 infectious disease threat to US military operations (3). In 2013, the Global Emerging Infections Surveillance branch, in collaboration with its worldwide network of partner laboratories and the Naval Health Research Center’s Operational Infectious Diseases Directorate, launched the Global Travelers’ Diarrhea (GTD) study to address issues posed to the US military by TD (4,5). This article describes the epidemiology of TD cases among US military populations and adult travelers during 2018–2023. In addition, this article characterizes coinfections, bacterial virulence factors, and pathogen factors relevant for medical countermeasure development.
Partner Institutions and Enrollment Sites
Partner institutions included the Walter Reed Armed Forces Research Institute of Medical Sciences Research Unit Nepal located in Kathmandu, Nepal, which had enrollment sites in Kathmandu and Pokhara, Nepal; US Naval Medical Research Unit EURAFCENT detachment in Cairo, Egypt, which had enrollment sites in South Sinai Governorate, Egypt, and Djibouti City, Djibouti; US Naval Medical Research Unit SOUTH located in Lima, Peru, which had enrollment sites in Cusco, Peru, and Comayagua, Honduras; and Walter Reed Army Institute of Research Europe-Middle East located in Tbilisi, Georgia, which had enrollment sites in Tbilisi, Batumi, and Gardabani, Georgia (Figure 1).
Participant Enrollment
Each partner institution was required to have institutional review board approval before collecting GTD Study–associated samples. Upon determination of eligibility, participants signed an informed consent agreement, except at the US Naval Medical Research Unit EURAFCENT; its local institutional review board did not require informed consent after January 2022. Study participation was voluntary. The study period was October 2018–April 2023.
Inclusion and Exclusion Criteria
The study inclusion criteria were adult travelers, >18 years of age, originating from the United States or any other upper-middle or high-income country (Appendix Table 1) (6), who had been in the enrollment country for <1 year, seeking healthcare services for acute GI illness. We defined illness as either acute diarrhea or acute gastroenteritis beginning >3 days after home departure. Adult travelers for this study included US military personnel, government employees, and citizens (e.g., nongovernmental organization workers, tourists, students, etc.). Patients were excluded from the study if they were experiencing chronic, persistent GI symptoms of >7 days before enrollment or noninfectious diarrhea or could not produce a fecal sample.
Case
The GTD study defined acute diarrhea as >3 loose/liquid feces, or >2 loose/liquid feces plus >2 additional GI symptoms. Case definitions for acute gastroenteritis were >3 vomiting episodes plus >1 additional GI symptom, or >2 vomiting episodes plus >2 additional GI symptoms occurring within the past 24 hours. Additional GI symptoms include diarrhea, vomiting, nausea, flatulence, cramping, muscle aches, headache, decreased urination, loss of appetite, bloating, abdominal pain, joint aches, malaise, fatigue, fever, or bloody feces.
Questionnaire
We assisted participants with completing a structured, standardized questionnaire. The questionnaire included questions about data variables describing demographics, symptoms, travel history, disposition, functional abilities, and treatment received.
Sample Receiving and Processing
Fecal specimens were stored as raw feces, in Cary-Blair (CB) medium, or feces in Cary-Blair with indicator (CBI) transport medium at 4°C for a maximum of 48 hours before transportation at 4°C to the GTD partner laboratory to perform testing. The partner laboratory assessed fecal specimens to verify appropriate temperature. Specimens that were frozen or room temperature were discarded and excluded from the study. Specimens received as raw feces were processed to a 20% weight/volume solution in phosphate-buffered saline before downstream testing, and specimens received in CB or CBI transport medium were directly used for downstream testing. Samples were either tested immediately upon arrival or frozen at –80°C and batch tested later.
Nucleic Acid Extraction and PCR
We extracted total nucleic acid from 20% fecal suspensions, fecal suspensions in CB or CBI, or boil prepped bacterial suspensions by using the QIAamp Viral RNA Mini kit (QIAGEN, https://www.qiagen.com). We used a total nucleic acid template for real-time PCR (rPCR) assays. We chose gene targets based on previously published literature (7–10).
We conducted real-time reverse transcription PCR (rRT-PCR) specific for norovirus genogroup I and II (GI and GII) by using the AgPath One Step RT-PCR kit (Ambion-Thermo Fisher Scientific, https://www.thermofisher.com) (7). We conducted rPCR specific for bacteria to detect the following organisms: E. coli pathotypes including enterotoxigenic E. coli (ETEC), enteroaggregative E. coli (EAEC), enteropathogenic E. coli (EPEC), shiga-like toxin–producing E. coli (STEC), and enteroinvasive E. coli (EIEC)/Shigella (8,9); Campylobacter jejuni, including subspecies jejuni and doylei (10); and Salmonella enterica ssp. enterica (10). We conducted rPCR specific for ETEC colonization factors on total nucleic acid extracted from isolated colonies (9). We used the PerfeCTa qPCR Tough Mix Kit (Quantabio, https://www.quantabio.com) for DNA amplification in all bacterial PCR assays. Primer and probe sequences, PCR conditions, and multiplex assay details are available (Appendix Tables 2, 3). We conducted all PCR reactions by using the Applied Biosystems 7500 Fast Dx or 7500 Fast Real-Time PCR instruments (Thermo Fisher Scientific, https://www.thermofisher.com).
Bacteriology
We subcultured samples that were positive for ETEC by rPCR onto MacConkey agar and incubated them at 35–37°C for 18–24 hours (11). A laboratory testing schematic is available (Appendix Figure 1).
Coinfection Analysis and Data Management
We analyzed coinfections by 4 major pathogen groups: norovirus (GI and GII), E. coli (EAEC, ETEC, EPEC, STEC, and EIEC/Shigella), Salmonella, and Campylobacter. We then cleaned the deidentified questionnaire and laboratory testing data by using Excel (Microsoft, https://www.microsoft.com) or Tableau Desktop version 2023.3 (Tableau, https://www.tableau.com). We merged questionnaire data and laboratory data in Excel by using participant identification numbers as the linking identifier.
During October 2018–April 2023, a total of 512 participants who met the acute diarrhea or acute gastroenteritis case definitions were enrolled in the GTD study in Honduras (21%), Peru (3%), Egypt (3%), Djibouti (39%), Nepal (26%), and Georgia (8%) (Table 1). The average participant age was 34 (SD 12) Years. Among participants, 35% were female, 59% male, and 6% unidentified sex. Participants were primarily born in North America (45%) or Europe (22%); however, that did not necessarily imply country of origin before travel and enrollment in the study. Most participants were US military service members (58%) or tourists (20%) (Table 1).
Across all sites, 403 (79%) of 512 samples tested positive for >1 pathogens, identifying a total of 867 pathogens (Table 2). Of the 403 positive samples, 79.7% were single infections, 19.6% were double infections, and 0.7% were triple infections. No samples were positive for all 4 pathogen groups (Table 2).
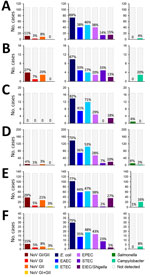
E. coli was the most common pathogen identified in Peru (67%), Nepal (77%), Georgia (75%), Honduras (69%), Egypt (82%), and Djibouti (70%), whereas Salmonella was the least identified in all countries except Egypt (6%) and Djibouti (6%) (Figure 2). Coinfection analysis identified E. coli in a higher number of coinfections than any of the other 3 pathogen groups across all 6 countries (Appendix Figure 2).
ETEC colonization factors were identified from ETEC isolates recovered from samples collected in 3 countries: Honduras, Djibouti, and Nepal (Table 3). In total, 106 isolates were tested. The most identified ETEC colonization factors were CS3 (25%) and CS21 (25%), as well as CS2 (18%) and CS6 (15%) (Table 3). The least identified ETEC colonization factors were CS17/19 (1%) and CS14 (2%). Colonization factors CS5 and CS7 were not identified from any country (Table 3).
This study describes the etiology and epidemiology of TD among US military and adult civilian travelers across South and Central America, Northern and sub-Saharan Africa, Southern Asia, and Eastern Europe. We found E. coli was the leading (67%–82%) etiology of TD across global surveillance sites (Figure 2). ETEC was the most identified E. coli pathotype in 5 of 6 countries (Figure 2). Our investigation also identified Campylobacter, Salmonella, and norovirus as TD etiologies, although with lower proportions than observed for E. coli (Figure 2). Those data on TD disease etiology are consistent with the literature discussing both military and civilian populations throughout the globe, indicating bacterial pathogens are the leading causative agents of disease (4,12–15). Our study data suggest E. coli, specifically pathotypes ETEC, EAEC, and EPEC, are the leading causes of TD in Southern and Central Asia, Northern Africa, the Middle East, sub-Saharan Africa, and Central and South America (Figure 2), which is consistent with the GTD study data for all 6 surveillance countries (12).
The highest rates of Campylobacter and Salmonella associated with TD are found in Southeast and East Asia, and high rates are also found in Southern and Central Asia (12). The GTD study did not include a surveillance site in Southeast or East Asia, but among included countries, we identified the highest rates of Campylobacter in Peru (20%) and Nepal (16%) and the highest rates of Salmonella in Egypt (6%) and Djibouti (6%) (Figure 2).
Previous work by the GTD study found E. coli (including all pathotypes tested for) in 42% of the cases enrolled during 2013–2018 (n = 410) (4), which is lower than the cases enrolled during 2018–2023 (n = 512; 72%) (Figure 2). That increase may represent improved laboratory diagnostic methods for E. coli pathotypes; samples collected during 2013–2018 were analyzed by conventional PCR, whereas samples collected during 2018–2023 were analyzed by rPCR. Of note, case numbers of Salmonella and Campylobacter were similar across study periods despite updates to laboratory protocols (Figure 2) (4).
Vaccines used prophylactically to prevent TD have the potential to reduce disease incidence and severity; however, no vaccines for E. coli, Campylobacter, or Shigella are currently licensed by the US Food and Drug Administration. ETEC vaccine candidates currently under investigation are based on antitoxin or anticolonization factor immunity. Approximately 50%–80% of all colonization factor-positive clinical ETEC isolates found within the general population encode colonization factors A/I, CS3, CS5, and CS6 (16), making them potential vaccine targets (17,18). In this study, CS6, CS3, CS2, and CS21 were the most identified colonization factors across the geographic regions tested; however, colonization factor A/I was only identified in 3% of isolates and CS5 was not identified in any isolate (Table 3). Our results combined with the efforts of the Global Emerging Infections Surveillance network of global surveillance laboratories, including maintaining repositories of clinical samples collected throughout the world, may help guide future vaccine development and therapeutics for TD and other diseases of interest to the US military and global health.
One limitation of this study is that we considered the disease etiology proportions from Egypt (n = 17), Peru (n = 15), and Georgia (n = 40) unstable because of low enrollment; we recommend interpreting the results with caution. In addition, selection bias may have influenced results because of site accessibility and the potential that travelers with mild TD are less likely to seek care than are travelers with severe TD, which might have affected the pathogens observed.
Moving forward, we recommend the GTD study expand to include antimicrobial resistance (AMR) characterization of bacterial pathogens identified from TD cases by using antimicrobial susceptibility testing and next-generation sequencing technologies to identify genetic markers of AMR and virulence factors of enteric bacterial pathogens. Those combined efforts could provide insight on the effects of AMR across unique global regions, enhance antimicrobial stewardship to limit changes in drug resistance patterns in enteric pathogens, and improve military health readiness through targeted prevention and treatment interventions for TD.
Dr. Anderson is the senior enteric infections scientist in the Operational Infectious Diseases Directorate at the Naval Health Research Center in San Diego, California, USA, contracted by General Dynamics Information Technology. Her research interests include infectious diseases, next-generation sequencing technologies for biosurveillance of enteric pathogens, and travelers’ diarrhea.
Acknowledgments
We thank Cailin La Clare, Reza Zeighami, Roger Pan, Paul Graf, Chris Myers, Amy Bogue, Tyler Moeller, Maria Silva, Maruja Bernal, Yoselinda Meza, Rina Meza, Jesus Rojas, Patricia Galvan, Roxana Caceda, Mayu Rivera, Faviola Reyes, Miguel Cabada Samame, Karen Mozo, Maria Luisa Morales, Martha Lopez, Eulogia Arque, Isabelle Nakhla, Edgie-Mark Co, Rebecca Pavlicek, Alexandria Kesterson, Ismail Raafat, the Camp Lemonnier, Djibouti Expeditionary Medical Facility team, Brian Walker, Christine Hulseberg, Thomas Musich, Shorena Mazmaniani, Tamar Jajanidze, Maia Mgeladze, Irma Kokaia, Inga Saghliani, Maia Samushia, Mikheil Patarkatsashvili, Shorena Mikava, Zhana Baramia, Walter Reed Research Unit Nepal staff, Walter Reed Army Institute of Research—Armed Forces Research Institute of Medical Sciences Department of Bacterial and Parasitic Diseases, Bhawani Thapa, Shristi Shakya, and Ananta Pokhrel for their work on the Global Travelers Diarrhea project, processing samples, and managing the project at the local level.
This work was supported by the Armed Forces Health Surveillance Division Global Emerging Infections Surveillance Branch (grant nos. P0120_23_SD, P0063_23_N6, P0135_23_N3, P0138_23_GA, and P0019_23_AF).
References
- Connor BA. Travelers’ diarrhea. In: Nemhauser J, editor. CDC yellow book 2024: health information for international travel [cited 2024 Jan 6]. Atlanta: Centers for Disease Control and Prevention; 2023. https://wwwnc.cdc.gov/travel/yellowbook/2024/preparing/travelers-diarrhea
- Steffen R. Epidemiology of traveler’s diarrhea. Clin Infect Dis. 2005;41(Suppl 8):S536–40. DOIPubMedGoogle Scholar
- US Army. Military infectious diseases research program (MIDRP) [cited 2024 Jan 6]. https://mrdc.health.mil/index.cfm/program_areas/medical_research_and_development/midrp_overview
- Ashbaugh HR, Early JM, Johnson ME, Simons MP, Graf PCF, Riddle MS, et al.; For The Gtd Study Team. A multisite network assessment of the epidemiology and etiology of acquired diarrhea among U.S. military and western travelers (global travelers’ diarrhea study): a principal role of norovirus among travelers with gastrointestinal illness. Am J Trop Med Hyg. 2020;103:1855–63. DOIPubMedGoogle Scholar
- Ashbaugh HR, Early JM, Johnson ME, Simons MP, Graf PCF, Riddle MS, et al. A prospective observational study describing severity of acquired diarrhea among U.S. military and Western travelers participating in the Global Travelers’ Diarrhea Study. Travel Med Infect Dis. 2021;43:
102139 . DOIPubMedGoogle Scholar - The World Bank. The world by region, 2019. [cited 2024 Jan 14]. https://datatopics.worldbank.org/world-development-indicators/the-world-by-income-and-region.html
- Chhabra P, Browne H, Huynh T, Diez-Valcarce M, Barclay L, Kosek MN, et al. Single-step RT-PCR assay for dual genotyping of GI and GII norovirus strains. J Clin Virol. 2021;134:
104689 . DOIPubMedGoogle Scholar - Hidaka A, Hokyo T, Arikawa K, Fujihara S, Ogasawara J, Hase A, et al. Multiplex real-time PCR for exhaustive detection of diarrhoeagenic Escherichia coli. J Appl Microbiol. 2009;106:410–20. DOIPubMedGoogle Scholar
- Liu J, Silapong S, Jeanwattanalert P, Lertsehtakarn P, Bodhidatta L, Swierczewski B, et al. Multiplex real time PCR panels to identify fourteen colonization factors of enterotoxigenic Escherichia coli (ETEC). PLoS One. 2017;12:
e0176882 . DOIPubMedGoogle Scholar - Wiemer D, Loderstaedt U, von Wulffen H, Priesnitz S, Fischer M, Tannich E, et al. Real-time multiplex PCR for simultaneous detection of Campylobacter jejuni, Salmonella, Shigella and Yersinia species in fecal samples. Int J Med Microbiol. 2011;301:577–84. DOIPubMedGoogle Scholar
- Versalovic J, Carroll KC, Funke G, Jorgensen JH, Landry ML, Warnock DW, editors. Manual of clinical microbiology, 11th ed. Washington: American Society for Microbiology Press; 2015.
- Lόpez-Vélez R, Lebens M, Bundy L, Barriga J, Steffen R. Bacterial travellers’ diarrhoea: A narrative review of literature published over the past 10 years. Travel Med Infect Dis. 2022;47:
102293 . DOIPubMedGoogle Scholar - Jennings MC, Tilley DH, Ballard SB, Villanueva M, Costa FM, Lopez M, et al. Case-case analysis using 7 years of travelers’ diarrhea surveillance data: preventive and travel medicine applications in Cusco, Peru. Am J Trop Med Hyg. 2017;96:1097–106. DOIPubMedGoogle Scholar
- Porter CK, Olson S, Hall A, Riddle MS. Travelers’ diarrhea: an update on the incidence, etiology, and risk in military deployments and similar travel populations. Mil Med. 2017;182(S2):4–10. DOIPubMedGoogle Scholar
- Walters WA, Reyes F, Soto GM, Reynolds ND, Fraser JA, Aviles R, et al. Epidemiology and associated microbiota changes in deployed military personnel at high risk of traveler’s diarrhea. PLoS One. 2020;15:
e0236703 . DOIPubMedGoogle Scholar - Nesta B, Pizza M. Vaccines against Escherichia coli. Curr Top Microbiol Immunol. 2018;416:213–42. DOIPubMedGoogle Scholar
- Pokharel P, Dhakal S, Dozois CM. The diversity of Escherichia coli pathotypes and vaccination strategies against this versatile bacterial pathogen. Microorganisms. 2023;11:344. DOIPubMedGoogle Scholar
- Rojas-Lopez M, Monterio R, Pizza M, Desvaux M, Rosini R. Intestinal pathogenic Escherichia coli: insights for vaccine development. Front Microbiol. 2018;9:440. DOIPubMedGoogle Scholar
Figures
Tables
Cite This ArticleOriginal Publication Date: November 11, 2024
Table of Contents – Volume 30, Supplement—October 2024
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Hunter Smith, Armed Forces Health Surveillance Division, 11800 Tech Rd, Suite 200, Silver Spring, MD 20904, USA
Top