Volume 30, Number 5—May 2024
Dispatch
Paranannizziopsis spp. Infection in Wild Vipers, Europe
Cite This Article
Citation for Media
Abstract
We describe the detection of Paranannizziopsis sp. fungus in a wild population of vipers in Europe. Fungal infections were severe, and 1 animal likely died from infection. Surveillance efforts are needed to better understand the threat of this pathogen to snake conservation.
Over the past few decades, fungal pathogens have been implicated in wildlife population declines, posing a substantial challenge to the conservation of many species, including herpetofauna (1). In reptiles, most fungal pathogens are within the genera Nannizziopsis, Paranannizziopsis, and Ophidiomyces, members of the order Onygenales (2). Of those genera, the most well-documented genus in wild reptiles is Ophidiomyces, consisting of the single species O. ophidiicola, which is responsible for ophidiomycosis, also called snake fungal disease (SFD) (3). Infections with Paranannizziopsis spp. fungi, on the other hand, are not well documented, possibly because of wide overlap with ophidiomycosis in how the disease manifests (4). Disease associated with Paranannizziopsis infection has been described in captive collections in North America (2,5–7) and Australasia (2,8). In wild populations, Paranannizziopsis spp. fungi have only been detected in nonnative free-living panther chameleons (Furcifer pardalis) from central Florida, USA (9), and in wild snakes in the United States and Canada (4). The geographic extent in wild host populations and severity of infection associated with Paranannizziopsis spp. fungi is unknown and deserves more thorough evaluation. We report infection with a Paranannizziopsis sp. fungus in 2 wild Seoane’s vipers (Vipera seoanei) from northwestern Spain. Handling of snakes was reviewed and approved by Virginia Tech Institute for Animal Care and Use Committee protocol 20-055. Vipers and tissue samples were collected under permit from Xunta de Galicia, Spain (permit no. EB-015/2021).
On May 14, 2021, two V. seoanei vipers, a subadult male (body length 31.7 cm, weight 10.3 g) and an adult female (body length 44.7 cm, weight 61 g), were captured near Zamáns in Vigo, Spain (42.16N, 8.68W; WGS1984). Both animals were in the process of molting and displayed many skin lesions on the head and body. The lesions were particularly abundant for the subadult male, for which the molting process was abnormal (i.e., dysecdysis). The animal was lethargic and appeared moribund. This snake was brought into captivity for supportive care but died the next day. The carcass was placed in ethanol until we performed necropsy and histopathological analyses. The adult female was reproductive, and, after we collected biometric data and skin swab samples, she was immediately released at the place of capture.
We swabbed the ventral and dorsal areas of the snakes in duplicate using a premoistened, sterile polyester-tipped applicator (Puritan, https://www.puritanmedproducts.com) and stored frozen swab samples at −20°C until analysis. We extracted DNA from the samples using PrepMan Ultra Sample Preparation Reagent (ThermoFisher Scientific, https://www.thermofisher.com). In addition, we excised 7 skin lesions (≈2 × 4 mm) from the subadult male at various locations across the body and stored them in 70% ethanol. We extracted DNA using a QIAGEN Blood and Tissue kit (QIAGEN, https://www.qiagen.com) following manufacturer’s instructions, which included a lyticase lysis step (200 U for 30 min at 30°C) to degrade fungal cell walls.
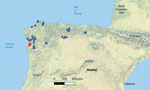
We screened extracted DNA from both the swab and tissue samples for the presence of Paranannizziopsis spp. and O. ophidiicola fungi using real-time PCR. O. ophidiicola fungi were not detected in any of the samples using a quantitative PCR (qPCR) targeting the internal transcribed spacer (ITS) region of the fungus (10). Samples from both vipers were qPCR-positive for Paranannizziopsis sp. fungus by genus-specific qPCR (4). We amplified and sequenced the full-length ITS and a portion of the mitochondrial cytochrome oxidase subunit III (COX3) gene (GenBank accession nos. OR353533 and OR351968) of the Paranannizziopsis sp. fungus, according to published methods (4). We compared sequences from those loci with existing sequences in GenBank using BLASTn (11). The ITS sequence most closely matched P. australasiensis strains in GenBank: 99.1%–99.6% identity over the ≈500 bp region sequenced for strains UAMH 12461 (OR100710), NWHC 24878–7 (OR100711), UAMH 12464 (OR100712), UAMH 12463 (OR100713), UAMH 10439 (KF477218), and UAMH 11645 (NR_111879). The COX3 sequence shared 99.9%–100.0% identity (over the ≈670-bp portion sequenced) with sequence data from P. australasiensis (strain UAMH 12461 [OR103159], NWHC 24878–7 [OR103160], UAMH 12464 [OR103161], UAMH 12463 [OR103162], UAMH 10439 [OR103163], UAMH 11645 [OR164]), P. californiensis (strain UAMH 10693 [OR103165]), and P. tardicrescens (strain CBS 142038 [OR103166]) in GenBank. After positive detections for Paranannizziopsis sp. fungi from those 2 animals, we also screened additional skin swab samples collected from V. seoanei vipers in Spain and Portugal in 2020 and 2021 (n = 37, including 1 with a ventral skin lesion). We did not identify Paranannizziopsis spp. fungi in those additional samples, indicating a pathogen prevalence of ≈5% (2 of 39 samples) (Figure 1).
At necropsy, the subadult male viper had variable numbers of multifocal to coalescing, raised, white-gray to dark brown discolored cutaneous lesions, ranging in size from 1 to 6 mm in diameter, along the left side of the mouth and labial scales; the lower jaw; and the central, dorsal, and caudal regions (Figure 2, panel A). We took samples of skin, bone, stomach, liver, kidney, and intestine for histopathological analysis. Tissue sections were stained by hematoxylin and eosin and Grocott-Gomori methenamine silver stains. Microscopically, skin lesions included areas of necrosis with granulocytic inflammation in the superficial to mid-epidermis; we observed slight edema adjacent to the mid-epidermis. Small chronic inflammatory cell aggregates composed of degenerated heterophils mixed with necrotic cellular debris and proteinaceous fluid were observed in these lesions. We detected nonpigmented fungal hyphae at the epidermal surface and breaching the epidermis under hematoxylin and eosin stain (Figure 2, panel B). We observed structures morphologically compatible with hyphae under Grocott-Gomori methenamine silver stain (Figure 2, panel C). Those hyphae were 1.8–4.9 µm in diameter, were septate, and had parallel walls with irregular dichotomous branching. We did not observe any relevant lesions or fungal elements in the internal tissues and viscera.
The effects of fungal diseases on reptiles have been difficult to evaluate, especially in cryptic species such as snakes. We report detection of a Paranannizziopsis sp. fungal infection in a wild population of V. seoanei vipers in Spain, and at least 1 viper likely died because of the infection. The pathology and fungal morphology were consistent with Paranannizziopsis spp. infections reported elsewhere (2,4–7). Although the strain detected in the snake that died was most similar to P. australasiensis, we were unable to identify the strain to a particular species of Paranannizziopsis fungus. Additional genetic analyses on the Paranannizziopsis sp. fungus detected in Spain might help better resolve its taxonomy.
Whether Paranannizziopsis spp. fungi are native to the Iberian Peninsula or whether our detections could represent recent transmission events from captive snakes remains unclear. We did not detect Paranannizziopsis spp. fungi in additional snakes sampled from Spain and Portugal, and O. ophidiicola fungus has not been detected in the Iberian Peninsula (11). Thus, if the pathogen was recently introduced, spread of this fungus to other vulnerable reptile populations is of concern, and further investigation is warranted considering the conservation need for most reptiles worldwide (12). V. seoanei vipers are a nearly endemic species to the Iberian Peninsula and is restricted to the northern region of the Atlantic climate (13). Populations have been severely impacted by habitat loss and fragmentation, and ecological models indicate high vulnerability of this species to climate change (14). Western populations, where these 2 infected snakes were found, are at the edge of the species’ distribution and are the most genetically diverse and isolated, highlighting their importance for maintaining genetic diversity (14,15). In light of this factor, detection of a species of Paranannizziopsis fungus raises concerns regarding the additive effects of other stressors and disease on the health of this imperiled population, and increased surveillance for this pathogen in wild populations might be warranted.
Ms. Blanvillain is a PhD candidate in the department of biological sciences at Virginia Tech. Her primary research interest is infectious disease in wildlife, specifically in herpetofauna.
Acknowledgments
We thank Megan Winzeler for laboratory assistance.
This material is based upon work supported by the NSF GRFP grant no. 480040. Additional funding to G.B. was provided by a Virginia Tech Cunningham fellowship, and a CeZAP (Center for Emerging, Zoonotic, and Arthropod-borne Pathogens) grant as part of the Infectious Diseases Interdisciplinary Graduate Education Program. F.M.-F. is supported by FCT - Fundação para a Ciência e a Tecnologia, Portugal (contract ref. DL57/2016/CP1440/CT0010).
References
- Fisher MC, Gurr SJ, Cuomo CA, Blehert DS, Jin H, Stukenbrock EH, et al. Threats posed by the fungal kingdom to humans, wildlife, and agriculture. MBio. 2020;11:e00449–20. DOIPubMedGoogle Scholar
- Sigler L, Hambleton S, Paré JA. Molecular characterization of reptile pathogens currently known as members of the chrysosporium anamorph of Nannizziopsis vriesii complex and relationship with some human-associated isolates. J Clin Microbiol. 2013;51:3338–57. DOIPubMedGoogle Scholar
- Lorch JM, Lankton J, Werner K, Falendysz EA, McCurley K, Blehert DS. Experimental infection of snakes with Ophidiomyces ophiodiicola causes pathological changes that typify snake fungal disease. MBio. 2015;6:e01534–15. DOIPubMedGoogle Scholar
- Lorch JM, Winzeler ME, Lankton JS, Raverty S, Snyman HN, Schwantje H, et al. Paranannizziopsis spp. infections in wild snakes and a qPCR assay for detection of the fungus. Front Microbiol. 2023;14:
1302586 . DOIPubMedGoogle Scholar - Bertelsen MF, Crawshaw GJ, Sigler L, Smith DA. Fatal cutaneous mycosis in tentacled snakes (Erpeton tentaculatum) caused by the Chrysosporium anamorph of Nannizziopsis vriesii. J Zoo Wildl Med. 2005;36:82–7. DOIPubMedGoogle Scholar
- Rainwater KL, Wiederhold NP, Sutton DA, Garner MM, Maguire C, Sanders C, et al. Novel Paranannizziopsis species in a Wagler’s viper (Tropidolaemus wagleri), tentacled snakes (Erpeton tentaculatum), and a rhinoceros snake (Rhynchophis boulengeri) in a zoological collection. Med Mycol. 2019;57:825–32. DOIPubMedGoogle Scholar
- Díaz-Delgado J, Marrow JC, Flanagan JP, Bauer KL, Zhang M, Rodrigues-Hoffmann A, et al. Outbreak of Paranannizziopsis australasiensis infection in captive African bush vipers (Atheris squamigera). J Comp Pathol. 2020;181:97–102. DOIPubMedGoogle Scholar
- Masters NJ, Alexander S, Jackson B, Sigler L, Chatterton J, Harvey C, et al. Dermatomycosis caused by Paranannizziopsis australasiensis in five tuatara (Sphenodon punctatus) and a coastal bearded dragon (Pogona barbata) in a zoological collection in New Zealand. N Z Vet J. 2016;64:301–7. DOIPubMedGoogle Scholar
- Claunch NM, Goodman CM, Harman M, Vilchez M, Smit SD, Kluever BM, et al. Dermatomycosis caused by Paranannizziopsis australasiensis in non-native panther chameleons (Furcifer pardalis) captured in Central Florida, USA. J Wildl Dis. 2023;59:322–31. DOIPubMedGoogle Scholar
- Bohuski E, Lorch JM, Griffin KM, Blehert DS. TaqMan real-time polymerase chain reaction for detection of Ophidiomyces ophiodiicola, the fungus associated with snake fungal disease. BMC Vet Res. 2015;11:95. DOIPubMedGoogle Scholar
- Blanvillain G, Lorch JM, Joudrier N, Bury S, Cuenot T, Franzen M, et al. Contribution of host species and pathogen clade to snake fungal disease hotspots in Europe. Commun Biol. 2024;7:440. DOIPubMedGoogle Scholar
- Cox N, Young BE, Bowles P, Fernandez M, Marin J, Rapacciuolo G, et al. A global reptile assessment highlights shared conservation needs of tetrapods. Nature. 2022;605:285–90. DOIPubMedGoogle Scholar
- Martínez-Freiría F, Brito JC. Vipera seoanei (Lataste, 1879). In: Milla AS, Ramos MA, editors. Reptiles [in Spanish]. 2nd ed. Fauna Ibérica, vol. 10. Madrid: Museo Nacional de Ciencias Naturales, CSIC; 2014. p. 942–56.
- Martínez-Freiría F. Assessing climate change vulnerability for the Iberian viper Vipera seoanei. Basic Appl Herpetol. 2015;29:61–80. DOIGoogle Scholar
- Martínez‐Freiría F, Velo‐Antón G, Brito JC. Trapped by climate: interglacial refuge and recent population expansion in the endemic Iberian adder Vipera seoanei. Divers Distrib. 2015;21:331–44. DOIGoogle Scholar
Figures
Cite This ArticleTable of Contents – Volume 30, Number 5—May 2024
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Gaëlle Blanvillain, Virginia Tech, 1015 Life Science Cir, Steger 352, Blacksburg, VA 24061, USA
Top