Volume 30, Number 6—June 2024
Dispatch
Concurrent Infection with Clade 2.3.4.4b Highly Pathogenic Avian Influenza H5N6 and H5N1 Viruses, South Korea, 2023
Figure 2
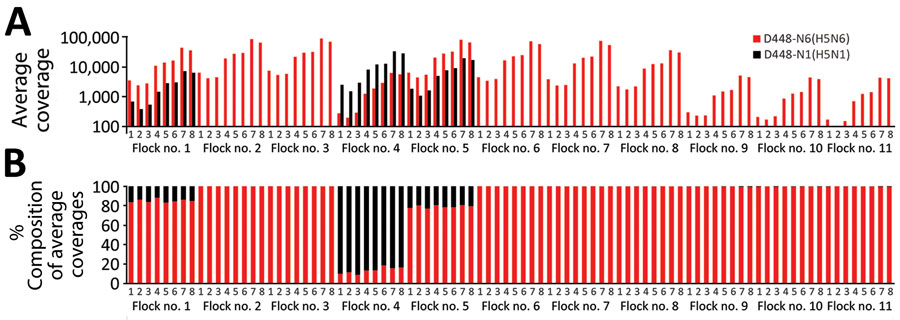
Figure 2. Co-infection status of birds at a broiler duck farm (D448) infected with clade 2.3.4.4b highly pathogenic avian influenza H5N6 and H5N1 viruses in South Korea, December 2023. Reference gene segments for mapping were designated as numbers 1–8; segment 1, polymerase basic 2; segment 2, polymerase basic 1; segment 3, polymerase acidic; segment 4, hemagglutinin; segment 5, nucleoprotein; segment 6, neuraminidase; segment 7, matrix; segment 8, nonstructural. A) Distribution of average coverages of the reads from each pooled oropharyngeal swab sample in the index farm, which had 11 flocks, mapped to the relevant reference viral genomes. Red bars indicate average mapping coverages of A/duck/Korea/D448-N6/2023(H5N6) to its reference gene segments and black bars indicate average mapping coverages of A/duck/Korea/D448-N1/2023(H5N1) to its reference gene segments. Three co-infected flocks (flocks 1, 4, and 5) had sufficient sequencing depth with >1,000-fold coverage of segments 4 (hemagglutinin) and 6 (neuraminidase). B) Distribution of percentage compositions of average coverages transformed from the average coverage values shown in panel A. Birds in flock 4 had more viral reads of H5N1 virus, whereas birds in flocks 1 and 5 had more reads of H5N6 virus. The other 8 flocks had the reads only mapped to H5N6 virus.