Volume 30, Number 8—August 2024
Dispatch
ST913-IVa-t991 Methicillin-Resistant Staphylococcus aureus among Pediatric Patients, Israel
Figure 4
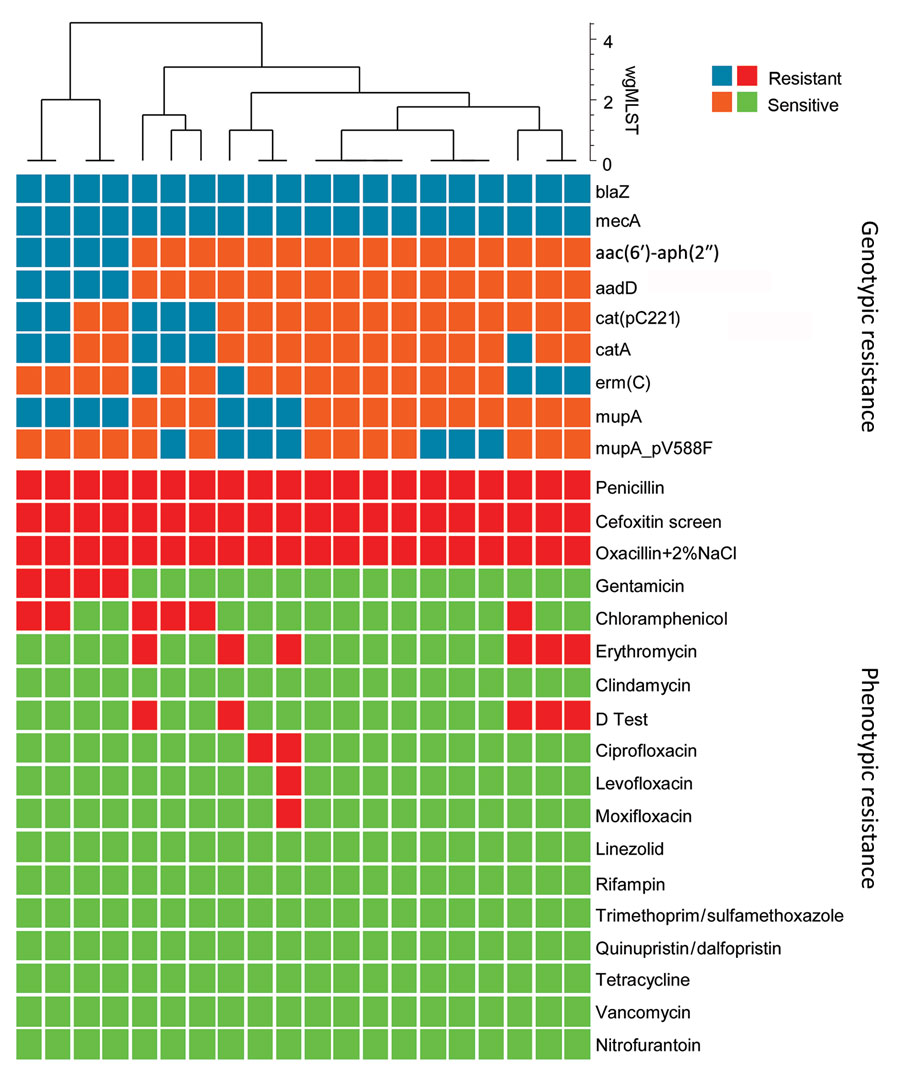
Figure 4. Comparison of genotypic and phenotypic resistance patterns of 20 t991 MRSA isolates from Israel tested using whole-genome sequencing. Blue tiles represent presence of resistance gene and orange tiles absence of resistance gene; red tiles represent antimicrobial resistance and green tiles antimicrobial sensitivity. Clustering is based on wgMLST data and generated by BioNumerics software (https://www.bionumerics.com). wgMLST, whole-genome multilocus sequence typing.
Page created: July 10, 2024
Page updated: July 21, 2024
Page reviewed: July 21, 2024
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.