Volume 30, Number 8—August 2024
Dispatch
ST913-IVa-t991 Methicillin-Resistant Staphylococcus aureus among Pediatric Patients, Israel
Cite This Article
Citation for Media
Abstract
In Israel, prevalence of sequence type 913, staphylococcal cassette chromosome mecIVa, spa type t991 methicillin-resistant Staphylococcus aureus lineage has surged among pediatric populations, predominantly in Arab and Orthodox Jewish communities. Antimicrobial resistance patterns vary by demographics. This lineage's spread and microevolution in the Middle East underscore the need for ongoing surveillance.
In 2010, a new methicillin-resistant Staphylococcus aureus (MRSA) clone, belonging to the clonal complex (CC) 913, Panton-Valentine leukocidin (PVL)–negative, staphylococcal cassette chromosome mec type IV, was isolated from Bedouin children in Israel (1). In 2012, isolates of CC913 were further analyzed, and their spa type was revealed as t991. Four t991 isolates were identified in hospitals across Israel, indicating the spread of the clone to communities beyond the Bedouin population in southern Israel (2). In 2015, a total of 12 t991 isolates were obtained from 280 patients (3), and in 2019, a total of 6 t991 isolates were obtained from 112 patients (4), mainly from children.
Since 2015, MRSA isolates of spa type t991 have emerged to become one of the main lineages in hospitals and health maintenance organizations in Israel. However, despite its significance, comprehensive characterization of spa type t991 clone is lacking. We explore its genomic context and antibiotic profile in this study.
During 2012–2020, the S. aureus national reference laboratory of Israel received a total of 4,646 MRSA isolates, obtained from skin and soft tissue infections (SSTIs), that were classified into 284 different spa types. Types t002, t008, and t032 were the most prevalent; t002 comprised 25% of total MRSA SSTI isolates, t008 comprised 15%, and t032, 5%. During that period, the proportion of t991 MRSA gradually increased to 13% of total MRSA SSTI isolates, whereas the leading spa types in MRSA SSTIs (t002 and t008) remained stable (Appendix Figure 1). During that period, 689 S. aureus samples of spa type t991, mecA-positive, PVL-negative, were received at the S. aureus national reference laboratory (Appendix). Most of the samples (406, 70%) were isolated from SSTIs; 66% isolates were from patients <5 years of age (p = 0.0001), whereas 4% were isolated from patients >60 years of age. In addition, most patients resided in localities associated with Arab and Orthodox Jewish populations (5). The number of t991 MRSA isolates from SSTI and blood increased dramatically, from 5 in 2012 to 180 in 2019 and 146 in 2020 (https://microreact.org/project/r4dFwJGXudh3gfyWtE1f87-t991final) (Figure 1).
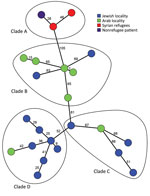
We conducted whole-genome sequencing on 20 t991 MRSA isolates that were selected (Appendix Table), along with 3 t991 MRSA isolates from Germany (6,7). The isolates clustered into 4 separate clades (Figure 2). Clade A consisted of the 3 t991 isolates from Germany and is 130 whole-genome multilocus sequence typing (wgMLST) alleles distant from the first isolate of clade B, which consisted of 7 isolates from patients who lived in the Negev and were admitted to the same hospital. Clade C consisted of 5 isolates from patients residing in the Jerusalem district. Clade D consisted of 8 isolates, 7 of which were from Orthodox Jewish patients.
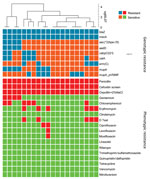
We tested 116 t991 MRSA isolates for phenotypic susceptibility by using the broth microdilution method (Figure 3). Isolates from patients living in Arab localities were more resistant to erythromycin and chloramphenicol, whereas those isolated from patients living in Jewish localities showed higher resistance to gentamicin, ciprofloxacin, levofloxacin, and moxifloxacin (Appendix Figure 2). That tendency was statistically significant for chloramphenicol (p = 0.01) and gentamicin (p = 0.01).
We found 9 antimicrobial resistance (AMR) determinants, 8 AMR genes and 1 point mutation, among the 20 t991 MRSA sequences (Figure 4). Overall, correlation between genotype prediction based on WGS and phenotypic AMR was 99% with a sensitivity of 94% and specificity of 100%. All discrepancies were associated with an absence of resistance determinant among phenotypically resistant isolates. No data for quinolone resistance genes or mutational resistance were predicted by the BioNumerics (https://www.bionumerics.com) or AMRFinder (https://github.com/ncbi/amr/releases/tag/amrfinder_v3.10.21) algorithms. In addition, we did not test phenotypic resistance against mupirocin.
We compared virulence profiles of 20 WGS t991 isolates with 3 t991 isolates from Germany, community-acquired MRSA USA300, and USA400 (8) as a reference using the functional genotyping tool of Bionumerics version 8.0. The main difference in virulence gene profile is reflected in the group of genes associated with adherence (Appendix Figure 3). In addition, virulence profiles can be grouped into 6 patterns on the basis of the presence of specific adherence factor genes in the genomes (Appendix Figure 4). We found no correlation between virulence profile and AMR, age, sex, residence location, or association with 1 of the genomic clades.
Next, to assess the genetic relationship of t991 isolates to other MRSA strains circulating worldwide, we created GrapeTree on the pubMLST site (https://pubmlst.org/organisms/staphylococcus-aureus) based on wgMLST data of representative local MRSA t991 strain (SA14675) along with 37,883 S. aureus global isolates (Appendix Figure 5) (9). The closest node to strain SA14675 is at a distance of 74 wgMLST alleles and is composed of 7 isolates. The next closest node is at a distance of 1,486 wgMLST alleles and composed of isolates that belong mainly to CC1.
Most t991 cases were isolated from young patients who live in strictly Orthodox and Arab localities. A possible explanation for this phenomenon is a similar lifestyle of the 2 sectors, characterized by overcrowding and large families. Regarding the evolution of this clone and its spread into the population, this strain appears to have evolved by multiple different genetic events. This assumption is supported by several findings. First, t991 MRSA isolates demonstrated classification into 4 distinct clades on the basis of geographic location and sectoral association; we noted genetic variation and weak clonality evident from the considerable distances between nodes, even within the same clade. Second, antibiotic resistance patterns vary between isolates obtained from patients who live in Jewish and Arab localities (Appendix Figure 2). Finally, spa type t991 composition is very short; it consists of 3 repeats (07-33-23) and can be formed as a result of genetic rearrangement of numerous MRSA strains harboring longer spa type repeats in which the repeats of spa type t991 from a part of their repeat succession. Worldwide phylogenetic analysis indicates that t991 MRSA stands out as a distinct emerging lineage because it appears considerably distant from most strains included in the GrapeTree (Appendix Figure 5).
Phenotypic AMR data for global isolates were available for 2 isolates (7,10). One isolate obtained in Kuwait was resistant to erythromycin, clindamycin, trimethoprim, and fusidic acid (10), and the other was isolated in Germany from a refugee from Syria (7) and was resistant to erythromycin, clindamycin, and tetracycline. Out of the 116 tested t991 isolates, none showed resistance to tetracycline by antimicrobial susceptibility. For 2 isolates, we observed phenotypic resistance for erythromycin and chloramphenicol without prediction of AMR determinant. Close inspection of those isolates revealed that were actually positive for ermC and cat, and their sequences were fragmented into multiple contigs. Consistent with previous publications (10–12), all isolates tested, except for the strain from the Syria refugee (7), were positive for eta, a toxin responsible for skin infections seen mainly among young patients (3,4,7). Those findings are in accordance with the observation that t991 MRSA is predominantly isolated from children.
In conclusion, our study shows the emergence of t991 MRSA in Israel. These strains affect mainly pediatric populations, and a geographic distribution is limited mainly to the Middle East. The epidemiologic and genomic information our research provides will assist further investigation on the origin and dissemination of this clone.
Dr. Baum is head of the National Staphylococcus Reference Laboratory at the Ministry of Health, Israel. His interests include genomic epidemiology and infectious diseases.
References
- Adler A, Givon-Lavi N, Moses AE, Block C, Dagan R. Carriage of community-associated methicillin-resistant Staphylococcus aureus in a cohort of infants in southern Israel: risk factors and molecular features. J Clin Microbiol. 2010;48:531–8. DOIPubMedGoogle Scholar
- Adler A, Chmelnitsky I, Shitrit P, Sprecher H, Navon-Venezia S, Embon A, et al. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in Israel: dissemination of global clones and unique features. J Clin Microbiol. 2012;50:134–7. DOIPubMedGoogle Scholar
- Biber A, Parizade M, Taran D, Jaber H, Berla E, Rubin C, et al. Molecular epidemiology of community-onset methicillin-resistant Staphylococcus aureus infections in Israel. Eur J Clin Microbiol Infect Dis. 2015;34:1603–13. DOIPubMedGoogle Scholar
- Hadyeh E, Azmi K, Seir RA, Abdellatief I, Abdeen Z. Molecular characterization of methicillin resistant Staphylococcus aureus in West Bank–Palestine. Front Public Health. 2019;7:130. DOIPubMedGoogle Scholar
- Shadmi E, Khatib M, Spitzer S. The COVID-19 Israeli tapestry: the intersectionality health equity challenge. Isr J Health Policy Res. 2023;12:17. DOIPubMedGoogle Scholar
- Kossow A, Stühmer B, Schaumburg F, Becker K, Glatz B, Möllers M, et al. High prevalence of MRSA and multi-resistant gram-negative bacteria in refugees admitted to the hospital-But no hint of transmission. PLoS One. 2018;13:
e0198103 . DOIPubMedGoogle Scholar - Creutz I, Busche T, Layer F, Bednarz H, Kalinowski J, Niehaus K. Evaluation of virulence potential of methicillin-sensitive and methicillin-resistant Staphylococcus aureus isolates from a German refugee cohort. Travel Med Infect Dis. 2022;45:
102204 . DOIPubMedGoogle Scholar - Thurlow LR, Joshi GS, Richardson AR. Virulence strategies of the dominant USA300 lineage of community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA). FEMS Immunol Med Microbiol. 2012;65:5–22. DOIPubMedGoogle Scholar
- Zhou Z, Alikhan NF, Sergeant MJ, Luhmann N, Vaz C, Francisco AP, et al. GrapeTree: visualization of core genomic relationships among 100,000 bacterial pathogens. Genome Res. 2018;28:1395–404. DOIPubMedGoogle Scholar
- Boswihi SS, Udo EE, Monecke S, Mathew B, Noronha B, Verghese T, et al. Emerging variants of methicillin-resistant Staphylococcus aureus genotypes in Kuwait hospitals. PLoS One. 2018;13:
e0195933 . DOIPubMedGoogle Scholar - Monecke S, Coombs G, Shore AC, Coleman DC, Akpaka P, Borg M, et al. A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant Staphylococcus aureus. PLoS One. 2011;6:
e17936 . DOIPubMedGoogle Scholar - Azarian T, Cella E, Baines SL, Shumaker MJ, Samel C, Jubair M, et al. Genomic epidemiology and global population structure of exfoliative toxin A–producing Staphylococcus aureus strains associated with staphylococcal scalded skin syndrome. Front Microbiol. 2021;12:
663831 . DOIPubMedGoogle Scholar
Figures
Cite This ArticleOriginal Publication Date: July 16, 2024
Table of Contents – Volume 30, Number 8—August 2024
| EID Search Options |
|---|
|
|
|
|
|
|


Please use the form below to submit correspondence to the authors or contact them at the following address:
Moti Baum, Ministry of Health Ya’akov Eliav 9, Jerusalem, Israel
Top