Volume 30, Number 8—August 2024
Online Report
Proposal for a Global Classification and Nomenclature System for A/H9 Influenza Viruses
Figure 2
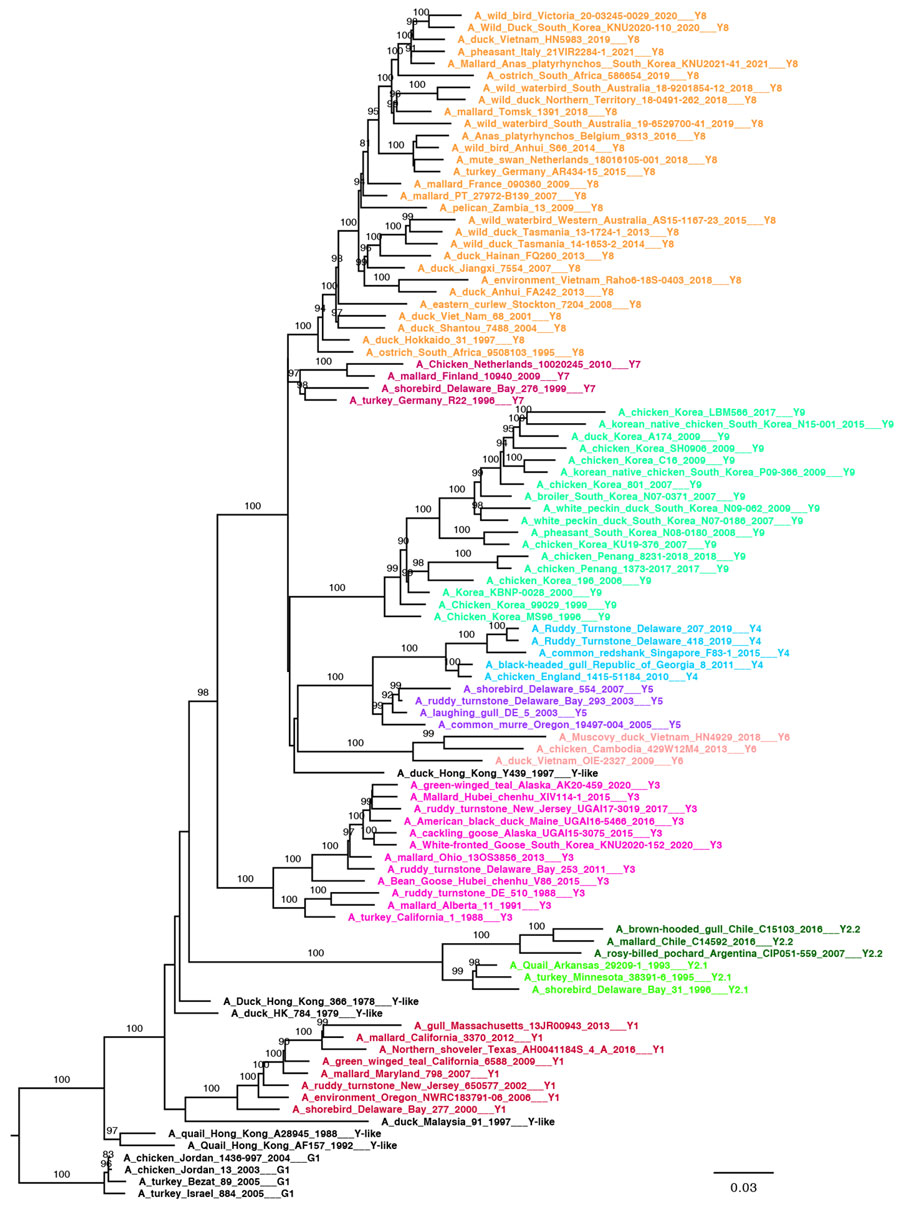
Figure 2. Pilot maximum-likelihood phylogenetic tree of the A/H9 influenza virus gene sequences obtained by using the representative dataset (Appendix 3) for the Y lineage provided as part of a proposed global classification and nomenclature system for A/H9 influenza viruses. Each clade is represented by >3 sequences, each labeled and colored according to the clade of belonging. Ultrafast-bootstrap supports >80% are indicated next to nodes. Scale bar indicates substitutions per site.
1These first authors contributed equally to this article.
2Members of The International H9 Evolution Consortium are listed at the end of this article.
Page created: July 12, 2024
Page updated: July 22, 2024
Page reviewed: July 22, 2024
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.