Volume 30, Number 8—August 2024
Dispatch
Characterization of Influenza D Virus Reassortant Strain in Swine from Mixed Pig and Beef Farm, France
Abstract
Influenza D virus was isolated from pigs on a mixed pig and beef farm in France. Investigation suggested bull-to-pig transmission and spread among pigs. The swine influenza D virus recovered was a reassortant of D/660 and D/OK lineages. Reported mutations in the receptor binding site might be related to swine host adaptation.
Influenza D virus (IDV) was initially isolated in 2011 from a pig with influenza-like symptoms (1). IDV has been identified globally in various species, including humans, without respiratory disease association according to serologic data (2,3) and sequencing from nasal washes (4). Cattle are the main IDV reservoir worldwide, including in France (5). Sow herds in France have tested positive for IDV antibodies, but IDV has not been isolated (6). We describe detection of IDV in fattening pigs in France and the accompanying genetic data for the swine origin IDV strain.
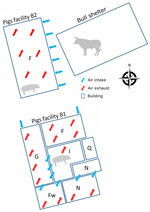
The sampled farm houses both pigs and cattle and is in the Brittany region of France. The pig herd operation is a farrow-to-finish system with 2 distinct barns: the first barn houses the sows and growing pigs in the nursery sectors and some of the fattening rooms (B1), and the other barn serves as the primary fattening barn (B2) (Figure 1). The pig herd had onset of chronic respiratory disorders that affected pigs in the nursery and fattening stages. The respiratory symptoms included sneezing and coughing.
A livestock veterinarian sampled tracheobronchial secretions from 7 fattening pigs (16 weeks of age) in October 2022. Pooled samples were submitted to the PathoSense laboratory (Ghent University, Merelbeke, Belgium) for sequencing (Oxford Nanopore Technologies, https://nanoporetech.com), which yielded results suggestive of an IDV infection. We then confirmed the presence of IDV genome in the tracheobronchial secretions by polymerase basic 1–gene qualitative reverse transcription PCR, as previously described (1). We successfully propagated strain D/swine/France/29-220655/2022 on swine testis cells. We confirmed viral growth by using polymerase basic 1–gene qualitative reverse transcription PCR and hemagglutination activity on chicken red blood cells (6).
We visited the farm in December 2022 to collect information about the layout and management. Our visit enabled us to assess the presence and spread of IDV in the pig barns. Because B2 had side air inlets close (7–10 m) to the bull shelter, we hypothesized that IDV was transmitted from the bovine reservoir to fattening pigs either through this air intake or from clothing and equipment used between both livestock sectors. We were unable to restrain the bulls for sampling, but the breeder emphasized the occurrence of respiratory signs, specifically coughing, in young bull calves. Those signs developed shortly after the arrival of the bull calves at 6 months of age. Because the calves originated from Pays de la Loire, the region with the highest serologic IDV prevalence in cattle and small ruminants in France (7), we considered a link between the signs and an IDV infection. We collected nasal swabs and blood samples from 30 pigs with influenza-like illness: 10 pigs from the nursery at 10 weeks of age (B1), 10 pigs from fattening at 15 weeks of age (B2), and 10 pigs from fattening at 25 weeks of age (B2). The 25-week-old pigs were from the same group as the 7 pigs sampled in October that were IDV positive. IDV genome was not detected in nasal swab samples. We tested for IDV antibodies in serum by using a hemagglutination inhibition assay and used the D/swine/France/29-220655/2022 strain as an antigen (8). The results indicated that the pigs exposed to IDV in October 2022 had seroconverted (Table), suggesting IDV spread within the group. In the nursery (B1), 10% of the serum was positive for IDV antibodies, indicating that IDV might also have circulated in the B1 facility among piglets or farrowing sows. The sows would have transmitted some passive immunity to piglets, and that immunity could be detected at the end of the postweaning period.
We sequenced the isolated D/swine/France/29-220655/2022 strain (Appendix) and conducted phylogenetic analysis. Our analysis revealed that the hemagglutinin-esterase fusion (HEF) sequence of D/swine/France/29-220655/2022 classified within the D/660 lineage. This lineage is closely related to contemporary bovine IDV strains in Italy but distantly related to recent bovine strains in France, which belong to the D/OK lineage (Appendix Figure). Whole-genome phylogeny also classified D/swine/France/29-220655/2022 within the D/660 lineage, again showing proximity to Italian bovine IDV strains (Figure 2, panel A). However, individual analysis of the 6 internal genomic segments revealed that 1 segment, the nucleoprotein-encoding gene, belonged to the D/OK lineage rather than D/660. This analysis indicated that D/swine/France/29-220655/2022 is a reassortant strain (Figure 2, panel B).
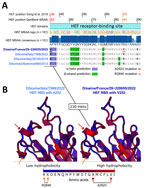
When aligned to the 151 publicly available IDV HEF amino acids sequences, D/swine/France/29-220655/2022 contained 2 unique mutations in the receptor binding site (RBS), A236V/A252V and R268K/R284K. We based amino-acid coordinates on previously published data (9) or GenBank multisequence alignment and translation to proteins (Figure 3, panel A). Those mutations are located close to the 230 helix and the 270 loop, which are essential structures forming the IDV HEF RBS open channel (9). Hydrophobicity is increased by the A252V mutation and can potentially change HEF RBS affinity to different receptors (Figure 3, panel B) without changing the global 3D structure of the protein.
In our study, we isolated a swine IDV strain in France, confirming serologic proof of IDV circulation in pig livestock (6). When considering the epidemiologic investigation of the mixed pig and beef farm, we believe there was a transmission of IDV from bulls to pigs. The D/swine/France/29-220655/2022 strain displayed an HEF associated with the D/660 clade. The D/660 clade has become increasingly prevalent in Europe since 2019, after a decline in the previously dominant D/OK clade (10–12). In addition, we found that D/swine/France/29-220655/2022 was a reassortant strain that included a nucleoprotein gene from the D/OK lineage. This reassortant strain likely emerged in bovine species before transmission to swine. Reassortment could have taken place in bulls housed in the assembly center before being dispatched to fattening farms, or in a farrowing farm that delivered animals to the study farm. Either case would imply co-circulation of D/660 and D/OK clades at some point in France, similar to other European countries (11,12). Because other reassortant strains belonging to D/660 but displaying D/OK genes have been isolated from cattle in Italy (10), the reassortant IDV might have been imported.
Of interest, D/swine/France/29-220655/2022 HEF exhibited 2 unique mutations (A252V and R284K) located in the RBS. The HEF receptor-binding cavity is known to form an open channel between the 230 helix and 270 loop that could be responsible for IDV broad-cell tropism (9). By increasing HEF RBS hydrophobicity, an A252V mutation might have promoted virus binding to pig cell receptors and subsequent uptake, demonstrating an adaptation to the pig host. Single-point mutations related to hydrophobicity changes, such as threonine to isoleucine, in RBS were shown to enable adaptation to new hosts in influenza C viruses, which also could be the case for IDV HEF (13). The reported mutations might have contributed to the spread of the virus among pigs within the fattening unit, as shown by the serologic data.
Our case shows that IDV can be transmitted from bovines to swine and adapt to its new host through potential specific mutations enhancing intraspecies transmission. This adaptation process is similar to findings from a previous study (14). Replicating such a pig–calves IDV transmission experiment by using a strain such as D/swine/France/29-220655/2022 could help validate our findings. The proliferation and spread of IDV in swine could be an issue for animal health. Surveillance for IDV should be incorporated with influenza A virus surveillance, particularly in mixed pig and beef herds or in pig herds situated near bovine livestock.
Dr. Richard is a researcher in molecular epidemiology at the swine virology immunology unit and National Reference Laboratory for swine influenza, France. His research interests include swine influenza virus evolution.
Acknowledgment
We thank the farm owner and the livestock veterinarian for their support of this study.
References
- Hause BM, Ducatez M, Collin EA, Ran Z, Liu R, Sheng Z, et al. Isolation of a novel swine influenza virus from Oklahoma in 2011 which is distantly related to human influenza C viruses. PLoS Pathog. 2013;9:
e1003176 . DOIPubMedGoogle Scholar - Trombetta CM, Montomoli E, Di Bartolo I, Ostanello F, Chiapponi C, Marchi S. Detection of antibodies against influenza D virus in swine veterinarians in Italy in 2004. J Med Virol. 2022;94:2855–9. DOIPubMedGoogle Scholar
- Trombetta CM, Marchi S, Manini I, Kistner O, Li F, Piu P, et al. Influenza D virus: serological evidence in the Italian population from 2005 to 2017. Viruses. 2019;12:30. DOIPubMedGoogle Scholar
- Leibler JH, Abdelgadir A, Seidel J, White RF, Johnson WE, Reynolds SJ, et al. Influenza D virus exposure among US cattle workers: A call for surveillance. Zoonoses Public Health. 2023;70:166–70. DOIPubMedGoogle Scholar
- Ducatez MF, Pelletier C, Meyer G. Influenza D virus in cattle, France, 2011-2014. Emerg Infect Dis. 2015;21:368–71. DOIPubMedGoogle Scholar
- Gorin S, Fablet C, Quéguiner S, Barbier N, Paboeuf F, Hervé S, et al. Assessment of influenza D virus in domestic pigs and wild boars in France: apparent limited spread within swine populations despite serological evidence of breeding sow exposure. Viruses. 2019;12:25. DOIPubMedGoogle Scholar
- Oliva J, Eichenbaum A, Belin J, Gaudino M, Guillotin J, Alzieu J-P, et al. Serological evidence of influenza D virus circulation among cattle and small ruminants in France. Viruses. 2019;11:516. DOIPubMedGoogle Scholar
- World Organisation for Animal Health. Influenza A viruses of swine. In: Manual of diagnostic tests and vaccines for terrestrial animals, 12th edition. Paris: The Organisation; 2023. p. 1–18.
- Song H, Qi J, Khedri Z, Diaz S, Yu H, Chen X, et al. An open receptor-binding cavity of hemagglutinin-esterase-fusion glycoprotein from newly-identified influenza D virus: basis for its broad cell tropism. PLoS Pathog. 2016;12:
e1005411 . DOIPubMedGoogle Scholar - Chiapponi C, Faccini S, Fusaro A, Moreno A, Prosperi A, Merenda M, et al. Detection of a new genetic cluster of influenza D virus in Italian cattle. Viruses. 2019;11:1110. DOIPubMedGoogle Scholar
- Gaudino M, Chiapponi C, Moreno A, Zohari S, O'Donovan T, Quinless E, et al. Evolutionary and temporal dynamics of emerging influenza D virus in Europe (2009–22). Virus Evol. 2022;8:081.
- Goecke NB, Liang Y, Otten ND, Hjulsager CK, Larsen LE. Characterization of influenza D virus in Danish calves. Viruses. 2022;14:423. DOIPubMedGoogle Scholar
- Szepanski S, Gross HJ, Brossmer R, Klenk HD, Herrler G. A single point mutation of the influenza C virus glycoprotein (HEF) changes the viral receptor-binding activity. Virology. 1992;188:85–92. DOIPubMedGoogle Scholar
- Kaplan BS, Falkenberg S, Dassanayake R, Neill J, Velayudhan B, Li F, et al. Virus strain influenced the interspecies transmission of influenza D virus between calves and pigs. Transbound Emerg Dis. 2021;68:3396–404. DOIPubMedGoogle Scholar
Figures
Table
Cite This ArticleOriginal Publication Date: July 19, 2024
1These first authors contributed equally to this article.
Table of Contents – Volume 30, Number 8—August 2024
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Gaëlle Simon, Laboratoire de Ploufragan-Plouzané-Niort, Rue des Fusillés, Zoopôle Les Croix, BP 53, 22440 Ploufragan, France
Top