Volume 30, Number 8—August 2024
Dispatch
Characterization of Influenza D Virus Reassortant Strain in Swine from Mixed Pig and Beef Farm, France
Figure 3
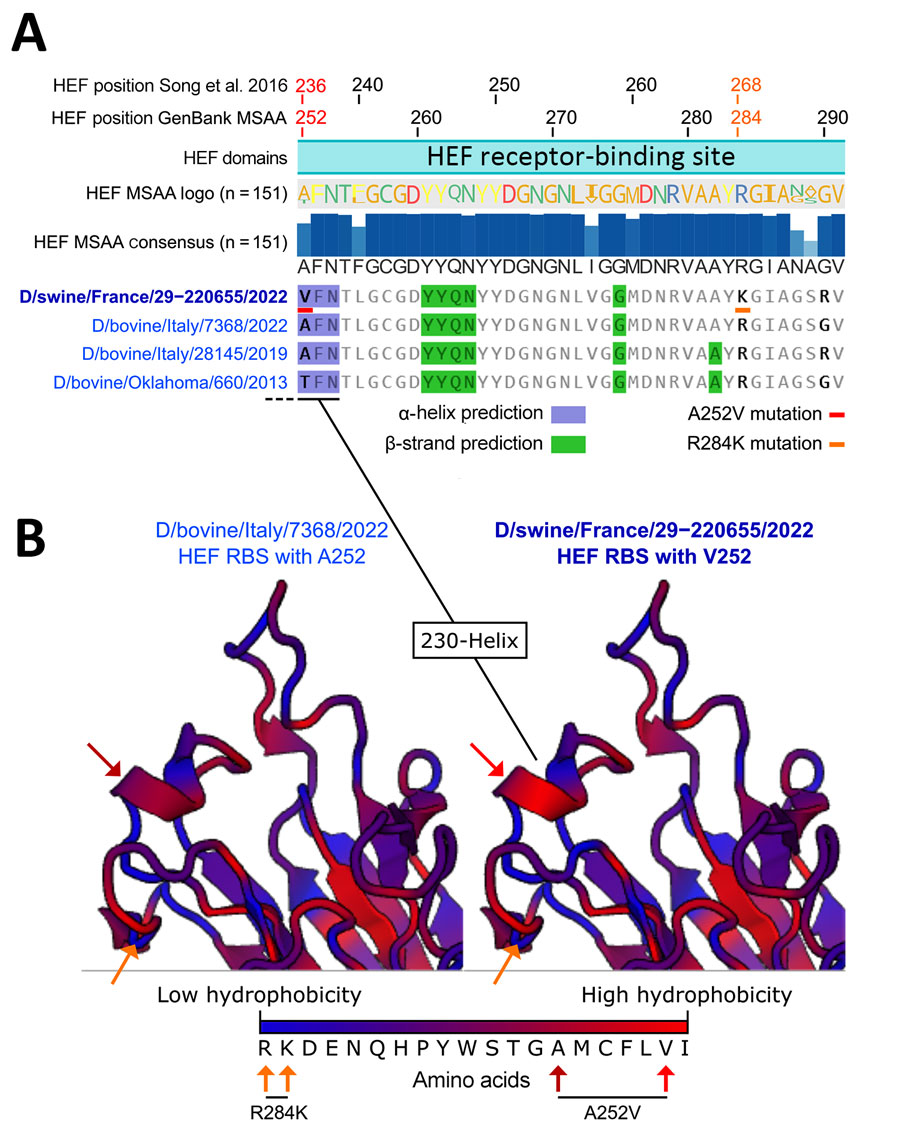
Figure 3. Alignments of hemagglutinin-esterase fusion protein sequences and structure prediction of the receptor-binding site of the influenza D virus strain recovered from swine in France. A) HEF sequence alignment and 2D structure prediction. From top to bottom: amino acid coordinates based on previously published research (9), amino acid coordinates based on GenBank MSAA and translation to protein, HEF domain based on previously published research (9), HEF sequence logo based on amino acid occurrence in the 151 HEF sequence alignment, HEF consensus sequence and percentage of amino occurrence in the 151 HEF sequence alignment, and representative HEF sequences from the D/660 lineage and their predicted secondary structures. Blue highlights indicate residues involved in α-helices and green highlights residues involved in β-strands. Red (A236V/A252V) and orange (R268K/R284K) underlines or arrows indicate unique mutations identified in D/swine/France/29-220655/2022. B) Predicted RBS structure of D/swine/France/29-220655/2022 compared with the closely related D/bovine/Italy/7368/2022. Blue colors on the protein 3D structures depict low hydrophobicity and red colors depict high hydrophobicity. HEF, hemagglutinin-esterase fusion; MSAA, multisequence alignment; RBS, receptor-binding site.
References
- Hause BM, Ducatez M, Collin EA, Ran Z, Liu R, Sheng Z, et al. Isolation of a novel swine influenza virus from Oklahoma in 2011 which is distantly related to human influenza C viruses. PLoS Pathog. 2013;9:
e1003176 . DOIPubMedGoogle Scholar - Trombetta CM, Montomoli E, Di Bartolo I, Ostanello F, Chiapponi C, Marchi S. Detection of antibodies against influenza D virus in swine veterinarians in Italy in 2004. J Med Virol. 2022;94:2855–9. DOIPubMedGoogle Scholar
- Trombetta CM, Marchi S, Manini I, Kistner O, Li F, Piu P, et al. Influenza D virus: serological evidence in the Italian population from 2005 to 2017. Viruses. 2019;12:30. DOIPubMedGoogle Scholar
- Leibler JH, Abdelgadir A, Seidel J, White RF, Johnson WE, Reynolds SJ, et al. Influenza D virus exposure among US cattle workers: A call for surveillance. Zoonoses Public Health. 2023;70:166–70. DOIPubMedGoogle Scholar
- Ducatez MF, Pelletier C, Meyer G. Influenza D virus in cattle, France, 2011-2014. Emerg Infect Dis. 2015;21:368–71. DOIPubMedGoogle Scholar
- Gorin S, Fablet C, Quéguiner S, Barbier N, Paboeuf F, Hervé S, et al. Assessment of influenza D virus in domestic pigs and wild boars in France: apparent limited spread within swine populations despite serological evidence of breeding sow exposure. Viruses. 2019;12:25. DOIPubMedGoogle Scholar
- Oliva J, Eichenbaum A, Belin J, Gaudino M, Guillotin J, Alzieu J-P, et al. Serological evidence of influenza D virus circulation among cattle and small ruminants in France. Viruses. 2019;11:516. DOIPubMedGoogle Scholar
- World Organisation for Animal Health. Influenza A viruses of swine. In: Manual of diagnostic tests and vaccines for terrestrial animals, 12th edition. Paris: The Organisation; 2023. p. 1–18.
- Song H, Qi J, Khedri Z, Diaz S, Yu H, Chen X, et al. An open receptor-binding cavity of hemagglutinin-esterase-fusion glycoprotein from newly-identified influenza D virus: basis for its broad cell tropism. PLoS Pathog. 2016;12:
e1005411 . DOIPubMedGoogle Scholar - Chiapponi C, Faccini S, Fusaro A, Moreno A, Prosperi A, Merenda M, et al. Detection of a new genetic cluster of influenza D virus in Italian cattle. Viruses. 2019;11:1110. DOIPubMedGoogle Scholar
- Gaudino M, Chiapponi C, Moreno A, Zohari S, O'Donovan T, Quinless E, et al. Evolutionary and temporal dynamics of emerging influenza D virus in Europe (2009–22). Virus Evol. 2022;8:081.
- Goecke NB, Liang Y, Otten ND, Hjulsager CK, Larsen LE. Characterization of influenza D virus in Danish calves. Viruses. 2022;14:423. DOIPubMedGoogle Scholar
- Szepanski S, Gross HJ, Brossmer R, Klenk HD, Herrler G. A single point mutation of the influenza C virus glycoprotein (HEF) changes the viral receptor-binding activity. Virology. 1992;188:85–92. DOIPubMedGoogle Scholar
- Kaplan BS, Falkenberg S, Dassanayake R, Neill J, Velayudhan B, Li F, et al. Virus strain influenced the interspecies transmission of influenza D virus between calves and pigs. Transbound Emerg Dis. 2021;68:3396–404. DOIPubMedGoogle Scholar
1These first authors contributed equally to this article.