Metagenomic Detection of Bacterial Zoonotic Pathogens among Febrile Patients, Tanzania, 2007–20091
Robert J. Rolfe, Sarah W. Sheldon, Luke C. Kingry, Jeannine M. Petersen, Venance P. Maro, Grace D. Kinabo, Wilbrod Saganda, Michael J. Maze, Jo E.B. Halliday, William L. Nicholson, Renee L. Galloway, Matthew P. Rubach, and John A. Crump

Author affiliations: Duke University Department of Medicine Division of Infectious Diseases and International Health, Durham, North Carolina, USA (R.J. Rolfe, M.P. Rubach, J.A. Crump); Centers for Disease Control and Prevention, Fort Collins, Colorado, USA (S.W. Sheldon, L.C. Kingry, J.M. Petersen); Kilimanjaro Christian Medical Centre, Moshi, Tanzania (V.P. Maro, G.D. Kinabo, M.P. Rubach, J.A. Crump); Kilimanjaro Christian Medical University College, Moshi (V.P. Maro, G.D. Kinabo, J.A. Crump); Mawenzi Regional Referral Hospital, Moshi (W. Saganda); University of Otago Department of Medicine, Christchurch, New Zealand (M.J. Maze); University of Glasgow School of Biodiversity, One Health and Veterinary Medicine, Glasgow, Scotland, UK (J.E.B. Halliday); Centers for Disease Control and Prevention, Atlanta, Georgia, USA (W.L. Nicholson, R.L. Galloway); Duke-National University of Singapore Programme in Emerging Infectious Diseases, Singapore (M.P. Rubach); Duke University Global Health Institute, Durham (M.P. Rubach, J.A. Crump); Centre for International Health, University of Otago, Dunedin, New Zealand (J.A. Crump)
Main Article
Figure 2
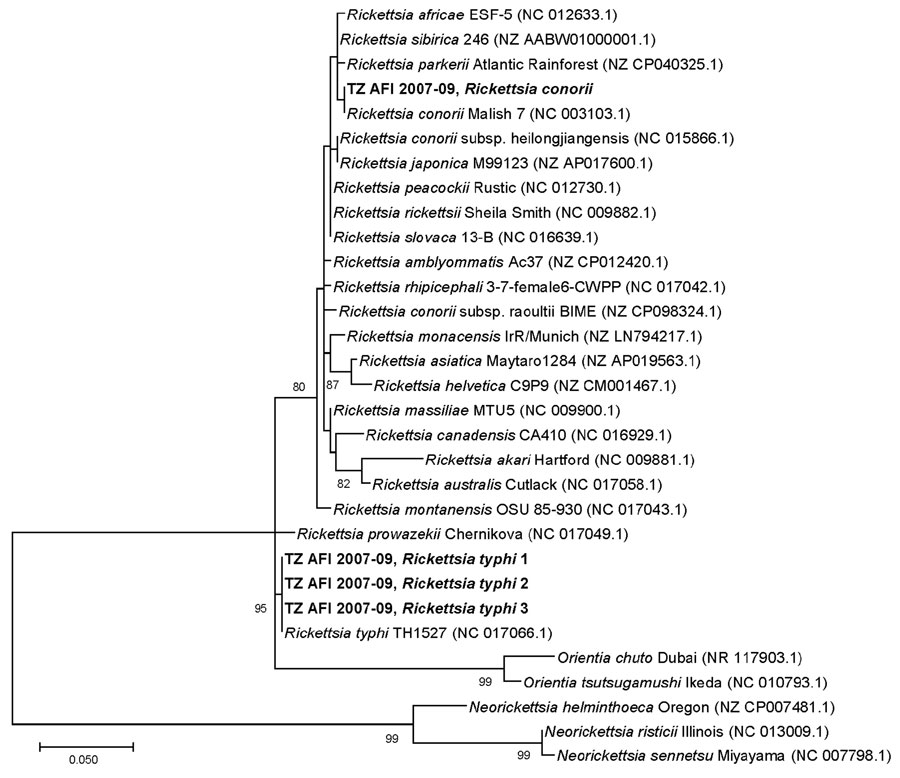
Figure 2. Phylogenetic tree of Rickettsia spp. sequences detected in metagenomic analysis of bacterial zoonotic pathogens among febrile patients, Tanzania, 2007–2009. The tree compares sequences from the 16S variable regions 1 and 2 (V1–V2) of the Rickettsia cohort from this study (bold text) to sequences from closely related Rickettsia species. Numbers in parentheses indicate GenBank accession numbers. The sequence from the study sample with R. conorii aligned 100% R. conorii strain Malish (accession no. NC003103.1) and was distinct from R. africae (accession no. NC012633.1). All 3 R. typhi strains from this study aligned 100% with R. typhi reference strain (accession no. NC017066.1) and were distinct from R. prowazekii (accession no. NC017049.1). The V1–V2 16S target is not sufficient to differentiate between R. conorii subsp. heilogjiangensis and R. japonica, or between R. rickettsia, R. peacockii, R. philipii, and R. slovaca. However, the V1–V2 16S target is sufficient to differentiate between R. conorii conorii and R. africae because 2 single-neucleotide differences would be expected between R. conorii conorii and R. africae and a TTT insertion in R. africae. Scale bar indicates nucleotide subsitutions per site.
Main Article
Page created: June 15, 2024
Page updated: July 20, 2024
Page reviewed: July 20, 2024
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
