Volume 30, Number 9—September 2024
Research
Autochthonous Leishmaniasis Caused by Leishmania tropica, Identified by Using Whole-Genome Sequencing, Sri Lanka
Figure 3
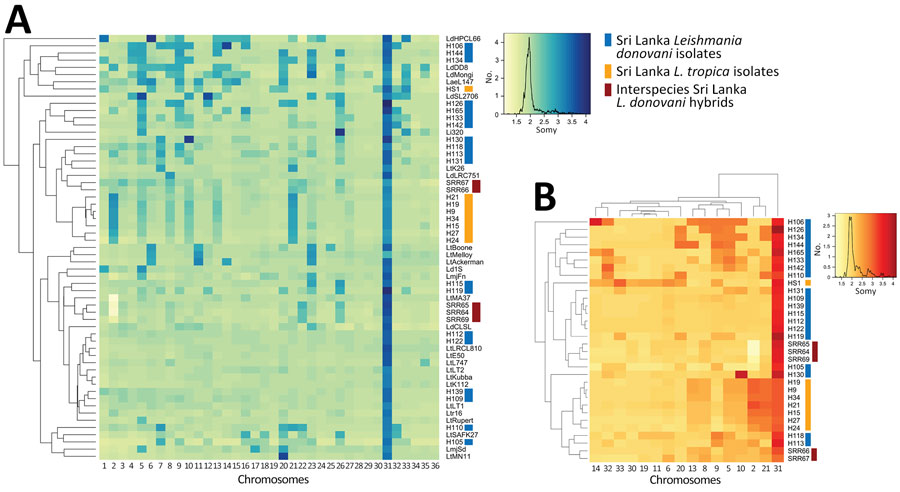
Figure 3. Highly conserved aneuploidy profiles within the cutaneous Leishmania tropica clade from Sri Lanka. A) Heat map visualization of Leishmania abnormal chromosome numbers in 27 patient isolates from Sri Lanka and 32 previously described strains or hybrids. L. tropica isolates from Sri Lanka are labeled with orange lines and L. donovani isolates from Sri Lanka are labeled with blue lines. B) Subset of the data shown in panel A highlighting only chromosomes that are polysomic in >1 isolate. Chromosome copy values were inferred from whole-genome analysis normalized read depth after mapping to the L. tropica L590 reference genome available on TriTrypDB (https://www.tritrypdb.org), with the assumption that all samples have a 2n DNA content. Somy, abnormal chromosome numbers.
1These authors contributed equally to this article.