Volume 30, Number 9—September 2024
Research
Autochthonous Leishmaniasis Caused by Leishmania tropica, Identified by Using Whole-Genome Sequencing, Sri Lanka
Figure 5
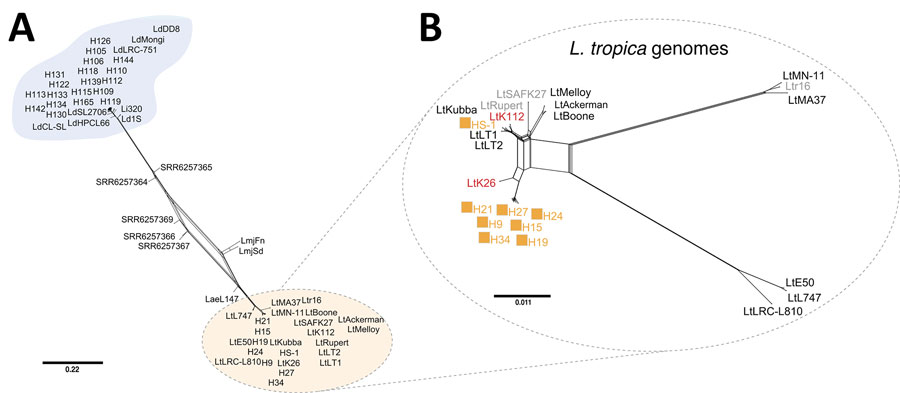
Figure 5. Phylogenetic network of all Leishmania isolates from Sri Lanka and Old World strains analyzed and visualized as a splits tree built using genomewide single-nucleotide polymorphisms in SplitsTree 6 (17). A) The cluster of the L. donovani complex is found at the top of the tree (blue) and the L. tropica are found at the bottom (orange). B) Phylogenetic network analysis of only the L. tropica genomes in our dataset. Orange squares, L. tropica isolates from Sri Lanka (HS1, H9–34); black font, Middle Eastern L. tropica; gray font, Azerbaijan (SAFK27) (8), Morocco (Ltr16) (21), and Afghanistan (Rupert) (8); red font, Indian L. tropica (K26 and K112) (8). Scale bar represents nucleotide substitutions per position.
References
- Desjeux P. Leishmaniasis: current situation and new perspectives. Comp Immunol Microbiol Infect Dis. 2004;27:305–18. DOIPubMedGoogle Scholar
- Alvar J, Yactayo S, Bern C. Leishmaniasis and poverty. Trends Parasitol. 2006;22:552–7. DOIPubMedGoogle Scholar
- Karunaweera ND, Pratlong F, Siriwardane HVYD, Ihalamulla RL, Dedet JP. Sri Lankan cutaneous leishmaniasis is caused by Leishmania donovani zymodeme MON-37. Trans R Soc Trop Med Hyg. 2003;97:380–1. DOIPubMedGoogle Scholar
- Siriwardana HVYD, Karunanayake P, Goonerathne L, Karunaweera ND. Emergence of visceral leishmaniasis in Sri Lanka: a newly established health threat. Pathog Glob Health. 2017;111:317–26. DOIPubMedGoogle Scholar
- Karunaweera ND, Senanayake S, Ginige S, Silva H, Manamperi N, Samaranayake N, et al. Spatiotemporal distribution of cutaneous leishmaniasis in Sri Lanka and future case burden estimates. PLoS Negl Trop Dis. 2021;15:
e0009346 . DOIPubMedGoogle Scholar - Zhang WW, Ramasamy G, McCall LI, Haydock A, Ranasinghe S, Abeygunasekara P, et al. Genetic analysis of Leishmania donovani tropism using a naturally attenuated cutaneous strain. PLoS Pathog. 2014;10:
e1004244 . DOIPubMedGoogle Scholar - Sterkers Y, Lachaud L, Crobu L, Bastien P, Pagès M. FISH analysis reveals aneuploidy and continual generation of chromosomal mosaicism in Leishmania major. Cell Microbiol. 2011;13:274–83. DOIPubMedGoogle Scholar
- Iantorno SA, Durrant C, Khan A, Sanders MJ, Beverley SM, Warren WC, et al. Gene expression in Leishmania is regulated predominantly by gene dosage. MBio. 2017;8:1–20. DOIPubMedGoogle Scholar
- Lypaczewski P, Thakur L, Jain A, Kumari S, Paulini K, Matlashewski G, et al. An intraspecies Leishmania donovani hybrid from the Indian subcontinent is associated with an atypical phenotype of cutaneous disease. iScience. 2022;25:
103802 . DOIPubMedGoogle Scholar - Inbar E, Shaik J, Iantorno SA, Romano A, Nzelu CO, Owens K, et al. Whole genome sequencing of experimental hybrids supports meiosis-like sexual recombination in Leishmania. PLoS Genet. 2019;15:
e1008042 . DOIPubMedGoogle Scholar - Karunaweera ND. Leishmania donovani causing cutaneous leishmaniasis in Sri Lanka: a wolf in sheep’s clothing? Trends Parasitol. 2009;25:458–63. DOIPubMedGoogle Scholar
- Lypaczewski P, Matlashewski G. Leishmania donovani hybridisation and introgression in nature: a comparative genomic investigation. Lancet Microbe. 2021;2:e250–8. DOIPubMedGoogle Scholar
- Zemanová E, Jirků M, Mauricio IL, Horák A, Miles MA, Lukeš J. The Leishmania donovani complex: genotypes of five metabolic enzymes (ICD, ME, MPI, G6PDH, and FH), new targets for multilocus sequence typing. Int J Parasitol. 2007;37:149–60. DOIPubMedGoogle Scholar
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–9. DOIPubMedGoogle Scholar
- Shaik JS, Dobson DE, Sacks DL, Beverley SM. Leishmania sexual reproductive strategies as resolved through computational methods designed for aneuploid genomes. Genes (Basel). 2021;12:1–16. DOIPubMedGoogle Scholar
- Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–67. DOIPubMedGoogle Scholar
- Salloum T, Moussa R, Rahy R, Al Deek J, Khalifeh I, El Hajj R, et al. Expanded genome-wide comparisons give novel insights into population structure and genetic heterogeneity of Leishmania tropica complex. PLoS Negl Trop Dis. 2020;14:
e0008684 . DOIPubMedGoogle Scholar - Dumetz F, Imamura H, Sanders M, Seblova V, Myskova J, Pescher P, et al. Modulation of aneuploidy in Leishmania donovani during adaptation to different in vitro and in vivo environments and its impact on gene expression. MBio. 2017;8:1–14. DOIPubMedGoogle Scholar
- Ferreira TR, Inbar E, Shaik J, Jeffrey BM, Ghosh K, Dobson DE, et al. Self-Hybridization in Leishmania major. MBio. 2022;13:
e0285822 . DOIPubMedGoogle Scholar - Bussotti G, Gouzelou E, Côrtes Boité M, Kherachi I, Harrat Z, Eddaikra N, et al. Leishmania genome dynamics during environmental adaptation reveal strain-specific differences in gene copy number variation, karyotype instability, and telomeric amplification. MBio. 2018;9:1–18. DOIPubMedGoogle Scholar
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–45. DOIPubMedGoogle Scholar
- Samarasinghe SR, Samaranayake N, Kariyawasam UL, Siriwardana YD, Imamura H, Karunaweera ND. Genomic insights into virulence mechanisms of Leishmania donovani: evidence from an atypical strain. BMC Genomics. 2018;19:843. DOIPubMedGoogle Scholar
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–6. DOIPubMedGoogle Scholar
- Shirian S, Oryan A, Hatam GR, Daneshbod Y. Three Leishmania/L. species—L. infantum, L. major, L. tropica—as causative agents of mucosal leishmaniasis in Iran. Pathog Glob Health. 2013;107:267–72. DOIPubMedGoogle Scholar
- Shirian S, Oryan A, Hatam GR, Daneshbod K, Daneshbod Y. Molecular diagnosis and species identification of mucosal leishmaniasis in Iran and correlation with cytological findings. Acta Cytol. 2012;56:304–9. DOIPubMedGoogle Scholar
- Bussotti G, Li B, Pescher P, Vojtkova B, Louradour I, Pruzinova K, et al. Leishmania allelic selection during experimental sand fly infection correlates with mutational signatures of oxidative DNA damage. Proc Natl Acad Sci U S A. 2023;120:
e2220828120 . DOIPubMedGoogle Scholar - Waitz Y, Paz S, Meir D, Malkinson D. Effects of land use type, spatial patterns and host presence on Leishmania tropica vectors activity. Parasit Vectors. 2019;12:320. DOIPubMedGoogle Scholar
- Sohrabi Y, Havelková H, Kobets T, Šíma M, Volkova V, Grekov I, et al. Mapping the genes for susceptibility and response to Leishmania tropica in mouse. PLoS Negl Trop Dis. 2013;7:
e2282 . DOIPubMedGoogle Scholar - Magill AJ, Grögl M, Gasser RA Jr, Sun W, Oster CN. Visceral infection caused by Leishmania tropica in veterans of Operation Desert Storm. N Engl J Med. 1993;328:1383–7. DOIPubMedGoogle Scholar
- Sacks DL, Kenney RT, Kreutzer RD, Jaffe CL, Gupta AK, Sharma MC, et al. Indian kala-azar caused by Leishmania tropica. Lancet. 1995;345:959–61. DOIPubMedGoogle Scholar
- Ghedin E, Zhang WW, Charest H, Sundar S, Kenney RT, Matlashewski G. Antibody response against a Leishmania donovani amastigote-stage-specific protein in patients with visceral leishmaniasis. Clin Diagn Lab Immunol. 1997;4:530–5. DOIPubMedGoogle Scholar
- Zhang WW, Matlashewski G. Characterization of the A2-A2rel gene cluster in Leishmania donovani: involvement of A2 in visceralization during infection. Mol Microbiol. 2001;39:935–48. DOIPubMedGoogle Scholar
- McCall LI, Matlashewski G. Localization and induction of the A2 virulence factor in Leishmania: evidence that A2 is a stress response protein. Mol Microbiol. 2010;77:518–30. DOIPubMedGoogle Scholar
- Al-Khalaifah HS. Major molecular factors related to Leishmania pathogenicity. Front Immunol. 2022;13:
847797 . DOIPubMedGoogle Scholar - Khan NH, Llewellyn MS, Schönian G, Sutherland CJ. Variability of cutaneous leishmaniasis lesions is not associated with genetic diversity of Leishmania tropica in Khyber Pakhtunkhwa province of Pakistan. Am J Trop Med Hyg. 2017;97:1489–97. DOIPubMedGoogle Scholar
- De Silva NL, De Silva VNH, Deerasinghe ATH, Rathnapala UL, Itoh M, Takagi H, et al. Development of a highly sensitive nested PCR and its application for the diagnosis of cutaneous leishmaniasis in Sri Lanka. Microorganisms. 2022;10:990. DOIPubMedGoogle Scholar
1These authors contributed equally to this article.