Volume 31, Number 10—October 2025
Research
Multidrug-Resistant pESI-Harboring Salmonella enterica Serovar Muenchen Sequence Type 82 in Poultry and Humans, Israel, 2020–2023
Figure 3
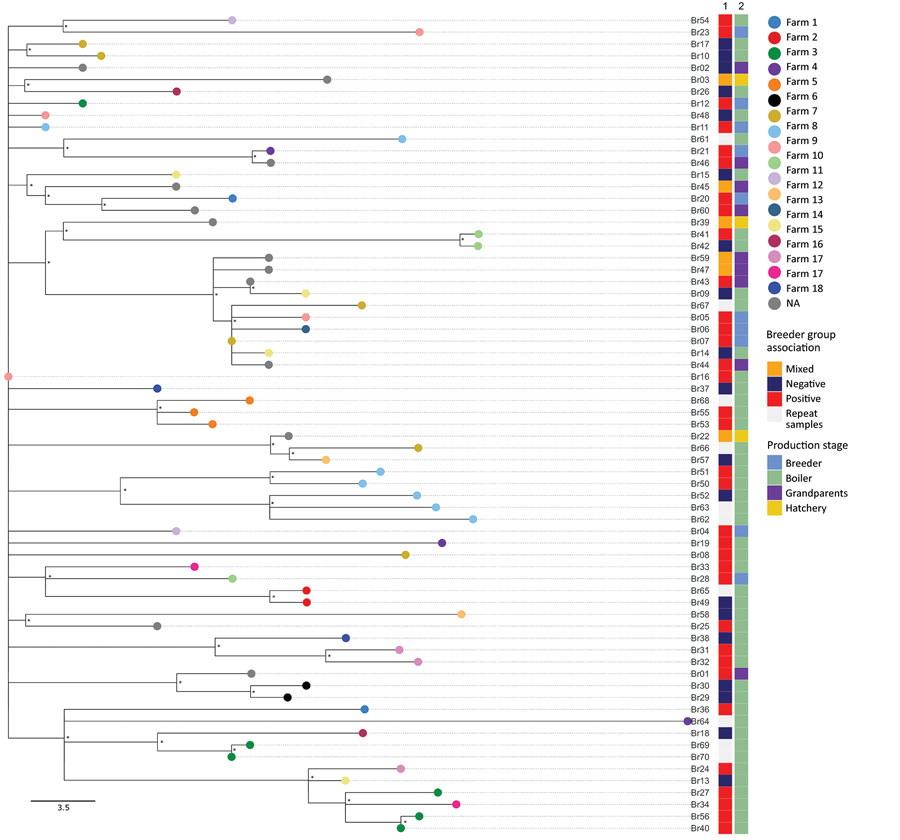
Figure 3. Maximum-likelihood tree reconstructed using whole-genome sequences of Salmonella enterica serovar Muenchen isolates collected from the various stages of the broiler production industry in study of Salmonella Muenchen in poultry and humans, Israel, June 2020–June 2023. We used a hybrid assembly of Salmonella Muenchen sequence type 82 (Br19) strain as a reference genome and used Salmonella Muenchen sequence type112 (National Center for Biotechnology Information Sequence Read Archive accession no. SRR6222324) to root the tree (not shown). The analysis included 874 single-nucleotide polymorphisms (SNPs) in the alignment (0–61 pairwise SNPs, median 30). Tip colors indicate the commercial broiler farm either where barns were tested (in the case of broiler samples), or the commercial broiler farm to which the breeder flock supplied (breeder samples). Asterisks (*) indicate nodes with >70% bootstrap support. Column 1 indicates data for the breeder group to which the broiler flocks were related (positive or control/negative). We labeled hatcheries and grandparent flocks as mixed if they received/supplied chicks from/to both positive and control heavy breeder farms. Column 2 indicates data for the production stage from which the sample was collected. Scale bar represents SNP difference.