Volume 31, Number 11—November 2025
Research
Isolation and Characterization of Rickettsia finnyi, Novel Pathogenic Spotted Fever Group Rickettsia in Dogs, United States
Cite This Article
Citation for Media
Abstract
In 2020, a novel spotted fever group Rickettsia was described in 3 clinically ill dogs in the United States. Using naturally infected canine blood, the novel Rickettsia sp. was isolated in epithelial (Vero E6) and mononuclear (DH82 and 030D) cell lines. The sequenced whole genome revealed a 1.27 Mb circular chromosome with 96.87% identity to Rickettsia raoultii on the basis of average nucleotide identity analysis. A maximum-likelihood phylogeny tree placed the novel Rickettsia in its own branch within the spotted fever group. Immunofluorescence revealed single rods localized along the membrane in epithelial cells and randomly distributed in the cytoplasm of mononuclear cells. We propose the name Rickettsia finnyi sp. nov., strain 2024-CO-Wats, which is available from national and international Rickettsial isolate reference collections. Fever and thrombocytopenia were among abnormalities in the 17 naturally infected dogs we describe, underscoring the pathogenic importance of R. finnyi sp. nov. and its potential public health relevance.
In 2020, a unique spotted fever group Rickettsia (SFGR), Rickettsia sp. 2019-CO-FNY, was identified in 3 clinically ill dogs in the southern and midwestern United States (1). Those dogs exhibited symptoms like those caused by R. rickettsii, the agent responsible for Rocky Mountain spotted fever (RMSF). SFGR are emerging tickborne pathogens infecting dogs and humans. Among tickborne pathogens infecting dogs, SFGR had the highest seroprevalence at 10.4% in the United States during 2004–2010 (2). The Centers for Disease Control and Prevention reported annual SFGR cases in humans in the United States increased substantially from 486 in 2000 to 6,248 in 2017 (3). Despite frequent exposure to SFGR, gaps remain in our understanding of pathogenic Rickettsia spp., disease severity, and tick vectors.
In the United States, several SFGR species, including R. parkeri, R. rickettsii, and R. rickettsii subsp. californica, cause disease in humans (4,5). Among those species, R. rickettsii is the most virulent in dogs and humans and can be fatal without early antibiotic intervention (6). In addition to R. rickettsii, other SFGR species have been detected in dogs in the United States, including R. montanensis, R. amblyommatis, and R. parkeri, all of which caused asymptomatic infection (7,8). Until recently, R. rickettsii was the only SFGR known to cause disease in dogs in North America. Dogs with RMSF can demonstrate fever, lethargy, neurologic signs, and generalized or localized pain, like arthropathy (9,10). Clinical signs reported in dogs infected with Rickettsia sp. 2019-CO-FNY resembled those seen in RMSF, indicating the existence of additional virulent SFGR in the United States and underscoring the importance of expanded vectorborne disease surveillance for canine and human health.
In this study, we cultured and sequenced a novel, pathogenic SFGR, Rickettsia sp. 2019-CO-FNY. We identified Rickettsia sp. 2019-CO-FNY in 14 additional sick dogs and cultured it from 1 infected dog. On the basis of whole-genome sequencing (WGS) and imaging, we determined that Rickettsia sp. 2019-CO-FNY is a new Rickettsia species, which we propose naming Rickettsia finnyi sp. nov., strain 2024-CO-Wats.
Infected Dogs
All dogs naturally infected with Rickettsia sp. 2024-CO-Wats were identified after samples were submitted to a veterinary diagnostic laboratory for canine comprehensive vectorborne disease testing. Signalment, sample collection date, and geographic location were included on submission forms. Attending veterinarians were asked to provide historical and clinical information. Ethical approval for animal use was not required for blood samples initially submitted for diagnostic testing; however, additional blood samples requested for R. rickettsii indirect immunofluorescence assay (IFA), posttreatment quantitative PCR (qPCR), and culture were approved under Institutional Animal Care and Use Committee protocol number 21–274. Analysis combined newly acquired data from 14 dogs with data from 3 dogs previously described (1).
Rickettsia Detection and Culture
The EDTA whole-blood sample used for culture was collected from a dog on April 25, 2024, by a veterinarian in Indiana, USA, and submitted to a veterinary diagnostic laboratory for comprehensive tickborne disease testing. The sample was received April 30, 2024, and stored at 4°C for 72 hours before testing. Tests consisted of qPCR for vertebrate GAPDH (internal control), Anaplasma, Apicomplexa, Babesia, Bartonella, Ehrlichia, hemotropic Mycoplasma and Rickettsia; IFA for Babesia vogeli, Bartonella henselae, Bartonella koehlerae, Bartonella vinsonii subsp. berkhoffii, Ehrlichia canis, and Rickettsii spp.; ELISA for antibodies to Babesia gibsoni and a SNAP 4DX Plus point-of-care ELISA (Idexx Laboratories, https://www.idexx.com) for Dirofilaria immitis antigen and species-specific antibodies to E. canis, Ehrlichia ewingii, Anaplasma phagocytophilum, A. platys, and Borrelia burgdorferi (11–17). Rickettsia sp. 2024-CO-Wats infection was confirmed with amplicon sequencing (GENEWIZ, http://www.genewiz.com) of the Rickettsia 23s-5s internal transcribed spacer genus qPCR and a newly developed R. finnyi species-specific (sp-sp) hydrolysis probe-based qPCR (1) (Appendix).
We added blood from a dog naturally infected with Rickettsia sp. 2024-CO-Wats to continuously maintained cell cultures using a previously published protocol (18). In brief, we combined 100 µL of blood and sucrose-phosphate-glutamate in a 1:1 ratio for each inoculation of 5 replicate cultures of Vero E6 (VE6) and 3 replicate cultures of DH82 and 030D cells seeded in either 7 ml tissue culture tubes, 6-well plates, or T-25 flasks (Fisher Scientific, http://www.fishersci.com) (Appendix Table 1). Cultures were grown at 34°C with 5% CO2 in either DMEM 5% FBS (VE6) or RPMI 1640 GlutaMAX 10% FBS (030D and DH82 cells) in a Biosafety Level 3 laboratory. We tested culture supernatants or cell suspensions from passages by qPCR and calculated fold changes in Rickettsia (Appendix). We performed retrospective qPCR and amplicon sequencing on stored culture DNA samples to assess a mutation acquired in a major facilitator superfamily (MFS) transporter gene (Appendix) (Table 1). We stained culture samples using the Gimenez method (19). We obtained images under oil immersion with an Olympus BX60 microscope and digital camera. We developed and performed an immunofluorescence technique on all 3 infected cell lines and acquired images with BZ-X810 Keyence (Appendix).
DNA Extraction and Whole-Genome Sequencing
We grew canine 030D and monkey VE6 cells infected with Rickettsia sp. 2024-CO-Wats in T25 flasks for DNA extraction (QIAGEN, https://www.qiagen.com). Sequencing was performed by the University of Delaware DNA Sequencing and Genotyping Center using Pacific Biosciences (https://www.pacb.com) Single-Molecule DNA for each culture DNA was sheared to 15 kb using the Megarupture 3 instrument (Diagenode, https://www.diagenode.com). SMRTbell DNA libraries were constructed according to the PacBio HiFi SMRTbell protocol using SMRTbell Express Template Prep Kit 3.0 and barcoded with the SMRTbell adaptor index plate 96A (Pacific Biosciences). AMPure PB beads were diluted at a 3.1× ratio to remove fragments <5 kb before sequencing and 1 SMRTcell was used to sequence the libraries on the Revio PacBio instrument for 30 hours.
Genome Assembly and Annotation
We assessed sequencing generated by PacBio HiFi circular consensus sequencing reads for quality using NanoPlot (20), aligned with host cell DNA, Canis lupus familiaris (030D) genomic DNA (Genbank no. GCA_000002285.4), and Chlorocebus sabaeus strain WHO RCB 10–87 (VE6) genomic DNA (Genbank no. GCA_015252025.1) using Minimap2 version 2.28 (21). We then filtered using SAMtools version 1.20 (22) and confirmed host DNA removal and coverage of the Rickettsia genome by using BAM file statistics. We assembled unmapped reads using Flye version 2.9.5 (23) and assessed quality using version 5.3 of the QUAST tool (24). We verified genome completeness and absence of plasmid sequences using Benchmarking Universal Single-Copy Orthologs tool version 5.8.0 and SourceFinder version 1.0 (25). We annotated the genome with the National Center for Biotechnology Information Prokaryotic Genome Annotation Pipeline.
Genomic and Phylogenetic Analysis
We directly compared the whole genomes of 37 Rickettsia spp. from GenBank, including 4 strains of R. rickettsii and 4 strains of R. parkeri, with the genome of 2024-CO-Wats (Appendix Table 2). We analyzed digital DNA-DNA hybridization (dDDH) with the Type Strain Genome Server (https://tygs.dsmz.de) (26) and determined average nucleotide identity (ANI) using the OrthoANI tool (27). We annotated 38 Rickettsia genomes with Prokka version 1.14.6 (default settings, Rickettsia-specific BLAST DB) (28). We identified orthologous core genes present in all genomes were identified using ProteinOrtho version 6.3.1 (29), aligned each gene at the nucleotide level with MAFFT version 7.526 (30), and concatenated into a matrix. We then performed maximum-likelihood phylogenetic inference with RAxML-NG version 1.2.1 (31) under a general time-reversible plus FC plus gamma 4m plus B model with per-partition parameter estimation. We mapped bootstrap support (500 replicates) onto the best maximum-likelihood tree, which was rooted with Rickettsia bellii.
Infected Dogs
We compiled signalment, collection date and location, and vectorborne diagnostic results from the 17 Rickettsia sp. 2024-CO-Wats infected dogs (14 new and 3 previously described) (Table 2) (1). More than half the samples were collected in May (9/17, 53%), and many dogs (7/17, 41%) resided in Kansas, Missouri, or Oklahoma (Table 2). Two dogs were co-infected with Babesia sp. (Coco) or Mycoplasma hematoparvum, and 5 dogs were seroreactive for other vectorborne pathogens. Because of limited and inconsistent clinical data, dogs were not uniformly evaluated for each parameter. The most common abnormal findings were fever (n = 13), lethargy (n = 13), and thrombocytopenia (n = 12) (Figure 1). Fourteen veterinarians administered doxycycline therapy (5–10 mg/kg every 12 hours) at the time of sample submission. One dog died before diagnosis and doxycycline therapy, and 1 was euthanized 1 day after starting doxycycline. One dog died because of nephrotic syndrome after treatment as previously described (1). Seven veterinarians sent additional samples for Rickettsia qPCR testing and R. rickettsii IFA. Results for all dogs tested for Rickettsia after doxycycline treatment were negative by qPCR, and all but 1 dog had a 4-fold or greater increase in R. rickettsii IFA titers (Table 2).
Rickettsia Culture and Visualization
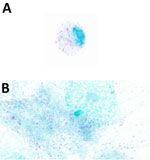
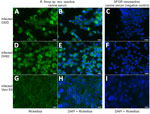
The EDTA-whole blood sample used for culturing was qPCR-positive for Rickettsia by 23s-5s ITS qPCR (quantification cycle 28.7, melting temperature 77°C) and R. finnyi–specific probe-based qPCR (quantification cycle 31) (Table 2). Serum from the same dog was seroreactive by R. rickettsii IFA with a titer of 1:2,048. All other vectorborne tests were negative (Table 2). All 3 cell lines were infected with Rickettsia sp. 2024-CO-Wats and maintained over multiple passages (Table 1; Appendix Figure 2). Images of 2024-CO-Wats-infected 030D-P18 and VE6-P4 cells stained with Gimenez revealed red, small (<0.5 by 2 µm), intracytoplasmic, randomly distributed rod-shaped bacteria (Figure 2). We visualized bacteria by immunofluorescence staining in 3 infected cell lines using serum from 2 dogs naturally infected with Rickettsia sp. 2024-CO-Wats (Figure 3). The staining did not differ between the 2 serum samples. The 030D-P9 (100% of cells), DH82-P8 (≈95%) and VE6-P2 (100%) cells were highly infected. The bacteria were in the cytoplasm as single rods or, less frequently, as clusters. In addition, we visualized aggregates of bacteria and, to a lesser extent, single bacilli on the cytoplasmic membrane of VE6 cells. We did not observe bacteria with nonreactive serum or with secondary antibody control.
Whole-Genome Sequencing, Assembly, and Annotation
We performed whole-genome sequencing using 1.33 µg (purity 1.92 A260/A280) of DNA from 030D cells infected with Rickettsia sp. 2024-CO-Wats and using 1.73 µg (purity 1.84 A260/A280) of DNA from VE6 cells infected with Rickettsia sp. 2024-CO-Wats. PacBio sequencing generated ≈1.5 million reads from infected 030D cells, where 1,481,022 reads were mapped to the host genome and removed leaving 16,333 unmapped reads. Approximately 4.6 million reads were generated from infected VE6 cells, where 3,030,649 reads were mapped to the host genome and removed, leaving 1,619,279 unmapped reads. We subsampled reads from VE6 cells and used 24,476 (2%) of the best quality reads for assembly. Flye generated a single, circularized contig of 1,270,764-bp with 32.3% G+C content for 2024-CO-Wats from both cultures. Mean genome coverage for 2024-CO-Wats was 174× from 030D and 261× from VE6 cultures. The assembled genomes showed 100% Benchmarking Universal Single-Copy Orthologs scores (genome completeness) using the Rickettsiales lineage. SourceFinder did not identify sequences originating from plasmids. Both genomes were deposited in Genbank as Rickettsia sp. 2024-CO-Wats cultured from 030D cells (accession no. CP170741) and Rickettsia sp. 2024-CO-Wats-2 from VE6 cells (accession no. CP187160). Both genomes were identical except a single nonsynonymous mutation at coordinate 646,406-bp (A/C) coding the 62nd amino acid in an MFS transporter protein (GenBank accession nos. XIA57199 and XRJ55031) changing the TTT (phenylalanine) in the 030D culture to TTG (leucine) in the VE6 culture. PCR and amplicon sequencing of the MFS transporter gene from cultures confirmed the mutation only occurred in VE6-P4 cells beginning on day 32 and was still present in VE6-P9 cells on day 104 (Table 1). The National Center for Biotechnology Information Prokaryotic Genome Annotation Pipeline identified 1,425 genes, 1,234 gene open-reading frames, 33 tRNAs, 3 rRNAs, and 4 ncRNAs.
Genomic and Phylogenetic Analysis
When compared with 37 Rickettsia spp. genomes, the 2024-CO-Wats genome was most similar to R. raoultii (GenBank accession no. CP098324); average nucleotide identity was 96.86% and dDDH was 70.6% (Appendix Table 2). ProteinOrtho identified 636 core orthologous genes, and phylogenetic analysis placed 2024-CO-Wats with its own distinct branch within the SFGR (Figure 4).
We isolated and sequenced the genome of a novel, pathogenic SFGR (formerly Rickettsia sp. 2019-CO-FNY) from a clinically ill dog that we propose naming Rickettsia finnyi sp. nov. type strain 2024-CO-Wats. This novel Rickettsia was cultured and maintained over many passages proving viability in epithelial (VE6) and mononuclear (030D and DH82) cells. Whole-genome sequencing generated a small, circular genome (1.27 kb) from infected 030D and VE6 cell lines. The Rickettsia genome was identical except for 1 nucleotide mutation in the VE6 cultured strain in an MFS transporter protein gene. The difference resulted in a conservative hydrophobic-to-hydrophobic amino acid change (phenylalanine to leucine), which could affect substrate specificity and transport efficiency, among other functions. The mutation could possibly have occurred in the VE6 culture because of different growth conditions or passage techniques. Genome alignment revealed it was most similar to R. raoultii (CP098324), with relatively small percentage differences between other SFGR species. Phylogenetic analysis of the genome sequence (CP170741) placed R. finnyi sp. nov. (2024-CO-Wats) on a distinct lineage within the spotted fever group, further supporting that it is a new species.
Criteria to designate a new Rickettsia sp. indicate that the genome must have an OrthoANI value of >83.63% compared with >1 Rickettsia species with a validly published name to be classified in the genus and a dDDH value of <92.3%, OrthoANI value of <99.19% identical with other known Rickettsia spp., or both to be considered a new species (32). R. finnyi sp. nov. (2024-CO-Wats) meets each of those criteria to be recognized as a new Rickettsia species. Genome comparisons revealed the highest results from both metrics was R. raoultii (CP098324) with a dDDH formula 2 value of 70.6% and OrthoANI measurement of 96.86%.
By both Gimenez staining and immunofluorescence, the morphology and cellular localization of R. finnyi sp. nov. (2024-CO-Wats) revealed characteristics consistent with pathogenic SFGR, including high quantities of bacillary-shaped intracellular bacteria and evidence of cell-to-cell expansion. In epithelial cells, R. finnyi sp. nov. (2024-CO-Wats) concentrated at the cytoplasmic membrane, likely representing direct transfer to neighboring cells, a well-documented mechanism used by SFGR for intracellular expansion (33). In contrast, the cytoplasmic localization observed in mononuclear cells is typical of obligate intracellular rickettsiae and provides supporting evidence that R. finnyi sp. nov. (2024-CO-Wats) is well adapted to survive in nonendothelial mammalian host cells (34).
In this study, we did not assess in vitro pathogenicity or cytopathic effects of R. finnyi sp. nov. (2024-CO-Wats); however, we documented growth for >104 days in 2 mononuclear cell lines. Previous studies have reported that pathogenic Rickettsia spp. can proliferate in nonendothelial cells, including leukocytes and macrophages, and that they exhibit enhanced intracellular survival in macrophage-like cells; the nonpathogenic Rickettsia spp. lacked this ability (34–38). Rickettsia spp. capable of proliferating in phagocytic cells have likely adapted mechanisms to evade host immunity and replicate before invading endothelial cells. The prolonged survival of R. finnyi sp. nov. (2024-CO-Wats) in mononuclear cells, along with clinical signs observed in naturally infected dogs, provide evidence that it is pathogenic. Additional studies comparing transcription levels and posttranslational modifications of R. finnyi sp. nov. in phagocytic versus epithelial cells might help elucidate mechanisms of pathogenicity and cytologic variation.
Since our initial description in 2020 of 3 clinically ill dogs naturally infected with R. finnyi sp. nov., an additional 14 infected dogs have been identified. Historical and clinicopathological findings for most of the dogs included a combination of fever, lethargy, and thrombocytopenia, like those seen in R. rickettsii infections. Vectorborne co-infections could have contributed to the abnormalities or disease severity in those dogs. However, the presence of similar abnormalities seen in the other dogs in our study without evidence of co-infections further supports the notion that R. finnyi sp. nov. is pathogenic in dogs.
Several Rickettsia spp., including R. parkeri and R. rickettsii, have been documented in both dogs and humans (6,8,39). Dogs serve as sentinels for human rickettsiosis, particularly RMSF, because they share similar clinical signs and exposure to the same ticks that transmit R. rickettsii (6,39,40). Antibodies to R. finnyi sp. nov. cross-react with R. rickettsii in IFA, as with most SFGR, making it challenging to accurately diagnose RMSF or other spotted fever rickettsiosises in dogs and humans. Furthermore, diagnostic PCRs specific to R. rickettsii might not detect novel Rickettsia spp. For example, Rickettsia sp. CA6269, which represents a novel Rickettsia sp. or subspecies of R. rickettsii, was detected in humans using broad-based Rickettsia qPCR screening after negative R. rickettsii and R. typhi sp-sp qPCR results (41). Studies are needed to determine whether R. finnyi sp. nov. can also infect and cause disease in humans.
R. finnyi sp. nov. (2024-CO-Wats) is likely transmitted by the lone star tick, Amblyomma americanum. Indeed, Noden et al. (42) reported amplified DNA sequences that were 100% identical with Rickettsia sp. 2019-CO-FNY in an A. americanum tick collected in 2018 in Oklahoma. Supporting possible exposure to A. americanum ticks, 1 R. finnyi sp. nov.–infected dog was co-infected with Babesia sp. coco, a protozoan pathogen detected in A. americanum ticks (43,44). Moreover, the geographic range of A. americanum ticks overlaps with areas in the United States where most infected dogs have been identified to date. Given the zoonotic potential of many Rickettsia spp., identifying the vectors and reservoir hosts of R. finnyi sp. nov. is essential toward understanding its transmission dynamics and potential public health impacts.
In conclusion, Rickettsia finnyi sp. nov. (fin′ny.i. N.L. gen. n. finnyi, named after Finny, the first infected dog, in recognition of companion dogs that have contributed to the discovery of novel pathogens) is proposed as a novel spotted fever group Rickettsia. Cells are small (<0.5 µm by 2 µm), rod-shaped intracytoplasmic bacteria that stain red using the Gimenez technique and grow in epithelial (Vero E6) and mononuclear (DH82 and 030D) cell lines. The circular genome is 1.27 Mb. This species has been identified in A. americanum ticks from Oklahoma and in dogs from the central and southeastern United States, where infection was associated with moderate to severe illness. The type strain is 2024-CO-Wats, isolated from a naturally infected dog in Tippecanoe County, Indiana, in 2024. Cultures have been deposited in 2 curated rickettsial banks: the Centers for Disease Control and Prevention Rickettsial Isolate Reference Collection (WDCM 1093; accession no. RFI001), Atlanta, Georgia, USA; and the Collection de Souches de l’Unité des Rickettsies (WDCM 875; accession no. R5053), Marseille, France.
Dr. Korla is a research associate at the North Carolina State College of Veterinary Medicine. His research interests include genome assembly and annotation, multi-omics data analysis and molecular modeling, and drug design. Mr. Karounos is a research specialist in the Department of Clinical Sciences at North Carolina State College of Veterinary Medicine. His research interests include acarology and bioinformatics.
Acknowledgments
We thank North Carolina State’s Vector-borne Disease Diagnostic Laboratory for use of equipment and assistance in sample processing; the University of Delaware DNA Sequencing and Genotyping Center for guidance and performance of whole genome sequencing; Joy A. Hecht and Christopher Paddock for assistance in depositing our cultures in the Rickettsial Isolate Reference Collection and assistance with sample shipments to France; Stephane Alibar and Pierre Edouard Fournier or assistance in depositing our cultures in France of the Collection de Souches de l’Unité des Rickettsies; Aharon Oren for assistance in Latin construction and review of the manuscript; and the attending veterinary clinicians and dog owners who sent additional samples for follow-up Rickettsia testing and provided medical and historical information about infected dogs.
This work was supported by American Kennel Club Canine Health Foundation (grant no. 02983).
B.A.Q., A.J.B., and E.E.B. provide continuing education on behalf of IDEXX Laboratories, Inc., and B.A.Q. receives partial salary support from IDEXX Laboratories, Inc. for veterinary medical applications. E.B.B. is the chief medical officer and J.M.W. is an employee of Galaxy Diagnostics. A.J.B. provides continuing education on behalf of Boehringer Ingelheim for veterinary medical applications.
During the preparation of this work, the authors used ChatGPT (https://chatgpt.com) to edit sentences for clarity and error resolution and debugging of bioinformatic workflows. After using ChatGPT, the authors reviewed and edited the content as needed and take full responsibility for the content of the publication.
References
- Wilson JM, Breitschwerdt EB, Juhasz NB, Marr HS, de Brito Galvão JF, Pratt CL, et al. Novel Rickettsia species infecting dogs, United States. Emerg Infect Dis. 2020;26:3011–5. DOIPubMedGoogle Scholar
- Yancey CB, Hegarty BC, Qurollo BA, Levy MG, Birkenheuer AJ, Weber DJ, et al. Regional seroreactivity and vector-borne disease co-exposures in dogs in the United States from 2004-2010: utility of canine surveillance. Vector Borne Zoonotic Dis. 2014;14:724–32.PubMedGoogle Scholar
- Centers for Disease Control and Prevention. Data and statistics on spotted fever rickettsiosis [cited 2025 Mar 25]. https://www.cdc.gov/rocky-mountain-spotted-fever/data-research/facts-stats/index.html
- Paddock CD, Karpathy SE, Henry A, Ryle L, Hecht JA, Hacker JK, et al. Rickettsia rickettsii subsp californica subsp nov, the Etiologic Agent of Pacific Coast Tick Fever. J Infect Dis. 2025;231:849–58.PubMedGoogle Scholar
- Rodino KG. Rickettsioses in the United States. Clin Microbiol Newsl. 2019;41:113–9. DOIGoogle Scholar
- Nicholson WL, Allen KE, McQuiston JH, Breitschwerdt EB, Little SE. The increasing recognition of rickettsial pathogens in dogs and people. Trends Parasitol. 2010;26:205–12. DOIPubMedGoogle Scholar
- Barrett A, Little SE, Shaw E. “Rickettsia amblyommii” and R. montanensis infection in dogs following natural exposure to ticks. Vector Borne Zoonotic Dis. 2014;14:20–5.PubMedGoogle Scholar
- Grasperge BJ, Wolfson W, Macaluso KR. Rickettsia parkeri infection in domestic dogs, Southern Louisiana, USA, 2011. Emerg Infect Dis. 2012;18:995–7. DOIPubMedGoogle Scholar
- Levin ML, Killmaster LF, Zemtsova GE, Ritter JM, Langham G. Clinical presentation, convalescence, and relapse of rocky mountain spotted fever in dogs experimentally infected via tick bite. PLoS One. 2014;9:
e115105 . DOIPubMedGoogle Scholar - Gasser AM, Birkenheuer AJ, Breitschwerdt EB. Canine Rocky Mountain Spotted fever: a retrospective study of 30 cases. J Am Anim Hosp Assoc. 2001;37:41–8. DOIPubMedGoogle Scholar
- Cerreta AJ, Yang TS, Ramsay EC, Birkenheuer AJ, Rahoi D, Qurollo B, et al. Detection of vector-borne infections in lions and tigers at two zoos in Tennessee and Oklahoma. J Zoo Wildl Med. 2022;53:50–9. DOIPubMedGoogle Scholar
- Ernst E, Qurollo B, Olech C, Breitschwerdt EB. Bartonella rochalimae, a newly recognized pathogen in dogs. J Vet Intern Med. 2020;34:1447–53. DOIPubMedGoogle Scholar
- Maggi RG, Birkenheuer AJ, Hegarty BC, Bradley JM, Levy MG, Breitschwerdt EB. Comparison of serological and molecular panels for diagnosis of vector-borne diseases in dogs. Parasit Vectors. 2014;7:127. DOIPubMedGoogle Scholar
- Hegarty BC, Qurollo BA, Thomas B, Park K, Chandrashekar R, Beall MJ, et al. Serological and molecular analysis of feline vector-borne anaplasmosis and ehrlichiosis using species-specific peptides and PCR. Parasit Vectors. 2015;8:320. DOIPubMedGoogle Scholar
- Qurollo BA, Archer NR, Schreeg ME, Marr HS, Birkenheuer AJ, Haney KN, et al. Improved molecular detection of Babesia infections in animals using a novel quantitative real-time PCR diagnostic assay targeting mitochondrial DNA. Parasit Vectors. 2017;10:128. DOIPubMedGoogle Scholar
- Beall MJ, Mainville CA, Arguello-Marin A, Clark G, Lemieux C, Saucier J, et al. An improved point-of-care ELISA for the diagnosis of anaplasmosis and ehrlichiosis during the acute phase of tick-borne infections in dogs. Top Companion Anim Med. 2022;51:
100735 . DOIPubMedGoogle Scholar - Birkenheuer AJ, Levy MG, Breitschwerdt EB. Development and evaluation of a seminested PCR for detection and differentiation of Babesia gibsoni (Asian genotype) and B. canis DNA in canine blood samples. J Clin Microbiol. 2003;41:4172–7. DOIPubMedGoogle Scholar
- Condit ME, Jones E, Biggerstaff BJ, Kato CY. Procedure for spotted fever group Rickettsia isolation from limited clinical blood specimens. PLoS Negl Trop Dis. 2022;16:
e0010781 . DOIPubMedGoogle Scholar - Ammerman NC, Beier-Sexton M, Azad AF. Laboratory maintenance of Rickettsia rickettsii. Curr Protoc Microbiol. 2008;Chapter 3:Unit 3A.5.
- De Coster W, Rademakers R. NanoPack2: population-scale evaluation of long-read sequencing data. Bioinformatics. 2023;39:btad311. DOIGoogle Scholar
- Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–100. DOIPubMedGoogle Scholar
- Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, et al. Twelve years of SAMtools and BCFtools. Gigascience. 2021;10:giab008. DOIGoogle Scholar
- Kolmogorov M, Yuan J, Lin Y, Pevzner PA. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol. 2019;37:540–6. DOIPubMedGoogle Scholar
- Mikheenko A, Saveliev V, Hirsch P, Gurevich A. WebQUAST: online evaluation of genome assemblies. Nucleic Acids Res. 2023;51(W1):W601–6. DOIPubMedGoogle Scholar
- Aytan-Aktug D, Grigorjev V, Szarvas J, Clausen PTLC, Munk P, Nguyen M, et al. SourceFinder: a machine-learning-based tool for identification of chromosomal, plasmid, and bacteriophage sequences from assemblies. Microbiol Spectr. 2022;10:
e0264122 . DOIPubMedGoogle Scholar - Meier-Kolthoff JP, Carbasse JS, Peinado-Olarte RL, Göker M. TYGS and LPSN: a database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res. 2022;50(D1):D801–7. DOIPubMedGoogle Scholar
- Lee I, Ouk Kim Y, Park S-C, Chun J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol. 2016;66:1100–3. DOIPubMedGoogle Scholar
- Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–9. DOIPubMedGoogle Scholar
- Klemm P, Stadler PF, Lechner M. Proteinortho6: pseudo-reciprocal best alignment heuristic for graph-based detection of (co-)orthologs. Front Bioinform. 2023;3:
1322477 . DOIPubMedGoogle Scholar - Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–80. DOIPubMedGoogle Scholar
- Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 2019;35:4453–5. DOIPubMedGoogle Scholar
- Diop A, El Karkouri K, Raoult D, Fournier P-E. Genome sequence-based criteria for demarcation and definition of species in the genus Rickettsia. Int J Syst Evol Microbiol. 2020;70:1738–50. DOIPubMedGoogle Scholar
- Teysseire N, Boudier JA, Raoult D. Rickettsia conorii entry into Vero cells. Infect Immun. 1995;63:366–74. DOIPubMedGoogle Scholar
- Curto P, Santa C, Allen P, Manadas B, Simões I, Martinez JJ. A Pathogen and a non-pathogen spotted fever group Rickettsia trigger differential proteome signatures in macrophages. Front Cell Infect Microbiol. 2019;9:43. DOIPubMedGoogle Scholar
- Kristof MN, Allen PE, Yutzy LD, Thibodaux B, Paddock CD, Martinez JJ. Significant growth by Rickettsia species within human macrophage-like cells is a phenotype correlated with the ability to cause disease in mammals. Pathogens. 2021;10:228. DOIPubMedGoogle Scholar
- Curto P, Simões I, Riley SP, Martinez JJ. Differences in intracellular fate of two spotted fever group Rickettsia in macrophage-like cells. Front Cell Infect Microbiol. 2016;6:80. DOIPubMedGoogle Scholar
- Voss OH, Gaytan H, Ullah S, Sadik M, Moin I, Rahman MS, et al. Autophagy facilitates intracellular survival of pathogenic rickettsiae in macrophages via evasion of autophagosomal maturation and reduction of microbicidal pro-inflammatory IL-1 cytokine responses. Microbiol Spectr. 2023;11:
e0279123 . DOIPubMedGoogle Scholar - Påhlson C, Lu X, Ott M, Nilsson K. Characteristics of in vitro infection of human monocytes, by Rickettsia helvetica. Microbes Infect. 2021;23:
104776 . DOIPubMedGoogle Scholar - Kidd L, Hegarty B, Sexton D, Breitschwerdt E. Molecular characterization of Rickettsia rickettsii infecting dogs and people in North Carolina. Ann N Y Acad Sci. 2006;1078:400–9. DOIPubMedGoogle Scholar
- Foley J, Backus L, López-Pérez AM. Focus on brown dog tick–transmitted Rocky Mountain spotted fever in dogs and people: shared threats and solutions [cited 2025 Apr 10]. https://avmajournals.avma.org/view/journals/javma/263/3/javma.24.11.0756.xml
- Probert WS, Haw MP, Nichol AC, Glaser CA, Park SY, Campbell LE, et al. Newly recognized spotted fever group Rickettsia as cause of severe Rocky Mountain spotted fever-like illness, northern California, USA. Emerg Infect Dis. 2024;30:1344–51. DOIPubMedGoogle Scholar
- Noden BH, Henriquez BE, Roselli MA, Loss SR. Use of an exclusion assay to detect novel rickettsiae in field collected Amblyomma americanum. Ticks Tick Borne Dis. 2022;13:
101959 . DOIPubMedGoogle Scholar - Bhosale CR, Wilson KN, Ledger KJ, White ZS, Dorleans R, De Jesus CE, et al. Ticks and tick-borne pathogens in recreational greenspaces in north central Florida, USA. Microorganisms. 2023;11:756. DOIPubMedGoogle Scholar
- Noden BH, Roselli MA, Loss SR. Effect of urbanization on presence, abundance, and coinfection of bacteria and protozoa in ticks in the US Great Plains. J Med Entomol. 2022;59:957–68. DOIPubMedGoogle Scholar
Figures
Tables
Cite This ArticleOriginal Publication Date: November 26, 2025
1These authors contributed equally to this article.
Table of Contents – Volume 31, Number 11—November 2025
| EID Search Options |
|---|
|
|
|
|
|
|


Please use the form below to submit correspondence to the authors or contact them at the following address:
Barbara A. Qurollo, North Carolina State University, College of Veterinary Medicine, Department of Clinical Sciences, Research Bldg, Office 464, 1060 William Moore Dr, Raleigh, NC 27606, USA
Top