Volume 31, Number 12—December 2025
Dispatch
Macrolide Resistance and P1 Cytadhesin Genotyping of Mycoplasma pneumoniae during Outbreak, Canada, 2024–2025
Abstract
We investigated macrolide resistance and P1 genotypes of Mycoplasma pneumoniae during the 2024–2025 outbreak in Hamilton, Ontario, Canada. Macrolide resistance remained stable at ≈10%–20%, but significant shifts in P1 genotype distribution and resistance rates in P1 types occurred, indicating notable changes in M. pneumoniae molecular epidemiology in Ontario since 2011–2012.
Mycoplasma pneumoniae is a cause of upper and lower respiratory tract infections, particularly in children, and occurs in endemic and epidemic patterns. Although tracheobronchitis is common, pneumonia is the most clinically significant manifestation, accounting for ≈4%–8% of community-acquired bacterial pneumonias during endemic periods (1). Macrolides remain the primary therapy for M. pneumoniae, but the global rise in clinically relevant resistance is posing increasing challenges to treatment (2). Since COVID-19 pandemic restrictions were scaled back during 2023, M. pneumoniae incidence and outbreaks have increased substantially worldwide (3–5). In Ontario, Canada, a delayed but unprecedented rise in detection, reaching up to 30% positivity, has been reported since May 2024 (6). We assessed macrolide resistance rates and P1 cytadhesin types of M. pneumoniae during the 2024–2025 outbreak in Hamilton, Ontario, Canada, and compared them with strains collected before the COVID-19 pandemic.
During January 2024–April 2025, a total of 4,297 nasopharyngeal swab (NPS) specimens from 3,717 patients were received by the Microbiology Laboratory of the Hamilton Regional Laboratory Medicine Program in Hamilton for M. pneumoniae detection by a laboratory-developed PCR. We screened all M. pneumoniae–positive samples (n = 417 after removing duplicates) for macrolide resistance by PCR genotyping and performed P1 cytadhesin typing on a randomly selected ≈25% subset of positive specimens from each month (n = 110) by amplifying the RepMP4 region of P1 cytadhesin gene (7) and sequencing with nanopore technology (Oxford Nanopore, https://nanoporetech.com) (Appendix). In addition to specimens received during the postpandemic period, additional M. pneumoniae–positive samples received during 2013–2020 were tested for macrolide resistance (n = 45) and P1 types (n = 23). We assessed statistical significance of differences in M. pneumoniae detection rates and macrolide resistance among different patient groups using the χ2 test with Yates correction to adjust for small sample size (Appendix).
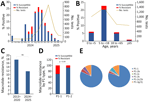
On average, 14.2% (381/2,680) of patients tested positive for M. pneumoniae in 2024, compared with 0.34% (2/576) in 2022 and 0.36% (2/555) in 2023. Since May 2024, the positivity rate gradually increased, reaching a peak of 22.5% in September 2024. After September, positivity rates steadily declined to <5% by January 2025, despite increased testing volumes through December 2024. Macrolide resistance rates varied by month and accounted for 11.8% of all positive samples during January 2024–April 2025; the highest rate of resistance (50%) was noted in July 2024 (Figure 1, panel A; Appendix Table 2). PCR genotyping identified only 1 single-nucleotide polymorphism (SNP) associated with macrolide resistance: A2063G, which is known to confer high-level macrolide resistance (erythromycin MIC >64 mg/L). This finding is consistent with a study reporting that >90% of isolates from Ontario during 2011–2012 carried the same mutation (8).
As expected, M. pneumoniae detection rates were much higher (≈20%) in children 5–<18 years of age than in other age groups. The macrolide resistance rate for M. pneumoniae in patients in this age group (≈11% of all positives) was not significantly different from the rates of resistance observed in children <5 years of age or adults 18–<65 years of age (Figure 1, panel B). In contrast, 50% of M. pneumoniae–positive strains from patients >65 years of age were macrolide resistant, a rate that was significantly higher than in children 5–<18 years of age (p = 0.02). Although the specimen number for the >65-year-old group was low (n = 6), the higher rate of macrolide resistance in this group could be related to higher likelihood of macrolide use in elderly patients.
We next compared macrolide resistance rates in M. pneumoniae strains during 2013–2020 and 2024–2025 (representing COVID-19 prepandemic and postpandemic periods), which were 17.8% (prepandemic) and 11.8% (postpandemic); the difference was not statistically significant (p = 0.24) (Figure 1, panel C). By P1 typing, 89/110 (81%) M. pneumoniae strains belonged to the P1-1 type, and 21/110 (19.1%) belonged to the P1-2 type. The macrolide resistance rate in P1-1–type M. pneumoniae strains was 29.9%, significantly higher (p = 0.04) than that in P1-2–type strains (7.7%) (Figure 1, panel D). During 2013–2020, 78.3% of M. pneumoniae strains were the P1-1 type, compared with 81% in 2024–2025. The proportions of P1-1 and P1-2 types were not significantly different between the 2 periods (p = 0.85). Among the P1-2 type M. pneumoniae strains, 2k, 2b, and 2g/2j variants were detected in specimens collected during 2013–2020. There was a predominance of 2g/2j variants (50%) in both periods, but the percentage of 2c/2k variants (36.4%) increased during 2024–2025 (Figure 1, panel E).
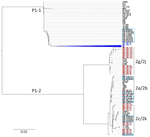
Phylogenetic analysis of the RepMP4 region of M. pneumoniae strains indicated that all P1-1 type strains (95.1%), including specimens from 2017–2020 (n = 16), clustered together on a distinct branch, separate from previously described strains from Ontario reported >10 years ago (8) (Figure 2). That previous study, conducted at the Public Health Ontario Laboratory, included representative specimens from across Ontario, including submissions from the Hamilton region (S.N. Patel, Public Health Ontario, pers. comm., email, 2025 Jul 28). Only 2 strains (MP_ON_05 and MP_ON_71) from the current outbreak showed homology with previously collected strains from Ontario, suggesting the RepMP4 region of P1-1–type strains has evolved over time while circulating in Ontario. Among the P1-2 types, 4 strains (15.4%) clustered with previously reported P1-2b variants, and 1 strain appeared related to P1-2c variants reported earlier in Ontario. In addition, our study revealed that P1-2 variants 2g/2j were circulating in Ontario as early as 2013, despite not having been previously reported in the region.
The first limitation of our study is that P1 typing was performed on only a subset of samples, and variant analysis relied solely on RepMP4 sequencing. Consequently, some P1-2 variants could only be assigned to variant groups (i.e., 2g/2j and 2c/2k). Moreover, few samples from 2013–2020 were available for P1 genotyping. Nevertheless, despite stable macrolide resistance rates, our findings show a major shift in the molecular epidemiology of M. pneumoniae since 2011-2012. Earlier Ontario data (2011–2012) reported P1-1 made up 38.1% of strains and P1-2 61.9% of strains (8), whereas in our study, 81% of P1-typed strains from the 2024–2025 outbreak were P1-1 type. Furthermore, unlike the previous study, which found no association between macrolide resistance and P1 types, our study shows significantly higher rates of macrolide resistance in the P1-1 group of M. pneumoniae. In addition, the distribution of P1-2 variants during the postpandemic period appeared more diverse than in the prepandemic period, and the P1-2c/2k variants expanded postpandemic. Our data, however, represent only the population of the Hamilton region; regional variation elsewhere in Ontario cannot be excluded.
The percentages of P1-1 versus P1-2 types in specimens from 2013–2020 (Figure 1, panel E) differs from previous reports for 2011–2012 (8). That discrepancy reflects that most prepandemic samples in our study (22/23) were collected during 2017–2020; only 1 was from 2013. Those data also suggest that the shift from predominantly P1-2 to P1-1 types might have begun before the pandemic.
Our study provides a snapshot of macrolide resistance rates and P1 genotypes of M. pneumoniae strains in Hamilton, Ontario, Canada, nearly a decade after the last provincial report. Macrolide resistance rates appear to have remained stable over that time, but we observed major changes in the P1 cytadhesin types of M. pneumoniae circulating in the Hamilton region. Clinicians and other public health professionals should be aware of those changes and their potential effects on clinical and public health management of respiratory infections caused by M. pneumoniae in Ontario.
Dr. Fatima is a postdoctoral fellow at the Research Institute of St. Joe’s and McMaster University. Her research focuses on genomic surveillance of infectious pathogens. Dr. Jayaratne is an associate professor at McMaster University. His primary research interest is the development of molecular diagnostic tests for infectious diseases.
Acknowledgments
We thank all laboratory technologists and technicians at the Virology and Molecular Microbiology Laboratories of the Hamilton Regional Laboratory Medicine Program for conducting routine testing for M. pneumoniae.
All sequences analyzed during this study have been submitted to GenBank (accession nos. PX307108–237).
References
- Waites KB, Xiao L, Liu Y, Balish MF, Atkinson TP. Mycoplasma pneumoniae from the respiratory tract and beyond. Clin Microbiol Rev. 2017;30:747–809. DOIPubMedGoogle Scholar
- Chen YC, Hsu WY, Chang TH. Macrolide-resistant Mycoplasma pneumoniae infections in pediatric community-acquired pneumonia. Emerg Infect Dis. 2020;26:1382–91. DOIPubMedGoogle Scholar
- Upadhyay P, Singh V. Mycoplasma pneumoniae outbreak in 2023: post-pandemic resurgence of an atypical bacterial pathogen. Cureus. 2024;16:
e58757 . DOIPubMedGoogle Scholar - Meyer Sauteur PM, Beeton ML; European Society of Clinical Microbiology and Infectious Diseases (ESCMID) Study Group for Mycoplasma and Chlamydia Infections (ESGMAC), and the ESGMAC Mycoplasma pneumoniae Surveillance (MAPS) study group. Mycoplasma pneumoniae: delayed re-emergence after COVID-19 pandemic restrictions. Lancet Microbe. 2024;5:e100–1. DOIPubMedGoogle Scholar
- Edouard S, Boughammoura H, Colson P, La Scola B, Fournier PE, Fenollar F. Large-scale outbreak of Mycoplasma pneumoniae infection, Marseille, France, 2023–2024. Emerg Infect Dis. 2024;30:1481–4. DOIPubMedGoogle Scholar
- Public Health Ontario. Mycoplasma pneumoniae testing at Public Health Ontario [cited 2025 Jun 11]. https://www.publichealthontario.ca/-/media/Documents/M/24/mycoplasma-pneumoniae-testing-pho.pdf
- Jiang FC, Wang RF, Chen P, Dong LY, Wang X, Song Q, et al. Genotype and mutation patterns of macrolide resistance genes of Mycoplasma pneumoniae from children with pneumonia in Qingdao, China, in 2019. J Glob Antimicrob Resist. 2021;27:273–8. DOIPubMedGoogle Scholar
- Eshaghi A, Memari N, Tang P, Olsha R, Farrell DJ, Low DE, et al. Macrolide-resistant Mycoplasma pneumoniae in humans, Ontario, Canada, 2010-2011. Emerg Infect Dis. 2013;19:1525–7. DOIPubMedGoogle Scholar
Figures
Cite This ArticleOriginal Publication Date: December 23, 2025
1These first authors contributed equally to this article.
Table of Contents – Volume 31, Number 12—December 2025
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Mohammad Rubayet Hasan, Hamilton Regional Laboratory Medicine Program, Department of Pathology and Molecular Medicine, McMaster University, Hamilton, ON L8N 4A6, Canada
Top