Volume 31, Supplement—May 2025
SUPPLEMENT ISSUE
Supplement
SARS-CoV-2 Genomic Surveillance from Community-Distributed Rapid Antigen Tests, Wisconsin, USA
Figure 4
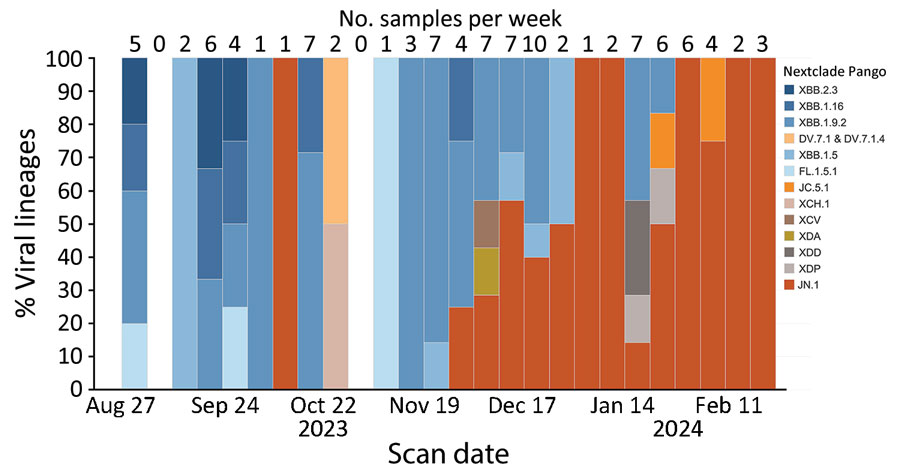
Figure 4. Number of samples collected per week and viral lineages detected in a study of SARS-CoV-2 genomic surveillance from community-distributed rapid antigen tests (RATs), Wisconsin, USA. The chart shows the percentage of SARS-CoV-2 lineages by week for samples that passed quality control thresholds of >90% of the SARS-CoV-2 genome at >10× depth. The date used reflected the date the participant scanned a provided QR code attached to a RAT. Unscanned RATs were excluded from the analysis. The number of samples included in each week’s percentage is shown above the bar. We assigned Pango lineages by using Nextclade version 3.5.0 (25). From August to mid-November 2023, the most common lineages in our samples fell under XBB.1.5, XBB.1.9.2, XBB.1.16, and XBB.2.3. Beginning in early December 2023, we began to see an increase in the number of samples belonging to the lineage JN.1, which dominated RAT samples scanned in February 2024.
References
- Ladner JT, Sahl JW. Towards a post-pandemic future for global pathogen genome sequencing. PLoS Biol. 2023;21:
e3002225 . DOIPubMedGoogle Scholar - Rasmussen M, Møller FT, Gunalan V, Baig S, Bennedbæk M, Christiansen LE, et al. First cases of SARS-CoV-2 BA.2.86 in Denmark, 2023. Euro Surveill. 2023;28:36. DOIPubMedGoogle Scholar
- Oliveira Roster KI, Kissler SM, Omoregie E, Wang JC, Amin H, Di Lonardo S, et al. Surveillance strategies for the detection of new pathogen variants across epidemiological contexts. PLOS Comput Biol. 2024;20:
e1012416 . DOIPubMedGoogle Scholar - Robishaw JD, Alter SM, Solano JJ, Shih RD, DeMets DL, Maki DG, et al. Genomic surveillance to combat COVID-19: challenges and opportunities. Lancet Microbe. 2021;2:e481–4. DOIPubMedGoogle Scholar
- World Health Organization. Recommendations for national SARS-CoV-2 testing strategies and diagnostic capacities [cited 2024 Apr 29]. https://www.who.int/publications/i/item/WHO-2019-nCoV-lab-testing-2021.1-eng
- Kates J, Cubanski J, Cox C, Published JT. Timeline of end dates for key health-related flexibilities provided through COVID-19 emergency declarations, legislation, and administrative actions [cited 2024 Nov 20]. https://www.kff.org/coronavirus-covid-19/issue-brief/timeline-of-end-dates-for-key-health-related-flexibilities-provided-through-covid-19-emergency-declarations-legislation-and-administrative-actions
- Centers for Disease Control and Prevention. COVID data tracker [cited 2024 May 9]. https://covid.cdc.gov/covid-data-tracker
- Centers for Disease Control and Prevention. COVID Museum COVID-19 timeline [cited 2025 Mar 6]. https://www.cdc.gov/museum/timeline/covid19.html
- Rader B, Gertz A, Iuliano AD, Gilmer M, Wronski L, Astley CM, et al. Use of at-home COVID-19 tests—United States, August 23, 2021–March 12, 2022. MMWR Morb Mortal Wkly Rep. 2022;71:489–94. DOIPubMedGoogle Scholar
- Khalid MF, Selvam K, Jeffry AJN, Salmi MF, Najib MA, Norhayati MN, et al. Performance of rapid antigen tests for COVID-19 diagnosis: a systematic review and meta-analysis. Diagnostics (Basel). 2022;12:110. DOIPubMedGoogle Scholar
- American Society for Microbiology. How the SARS-CoV-2 EUA antigen tests work [cited 2024 Jul 25]. https://asm.org:443/Articles/2020/August/How-the-SARS-CoV-2-EUA-Antigen-Tests-Work
- Food and Drug Administration. At-home OTC COVID-19 diagnostic tests [cited 2024 Jul 16]. https://www.fda.gov/medical-devices/coronavirus-covid-19-and-medical-devices/home-otc-covid-19-diagnostic-tests
- Martin GE, Taiaroa G, Taouk ML, Savic I, O’Keefe J, Quach R, et al. Maintaining genomic surveillance using whole-genome sequencing of SARS-CoV-2 from rapid antigen test devices. Lancet Infect Dis. 2022;22:1417–8. DOIPubMedGoogle Scholar
- Rector A, Bloemen M, Schiettekatte G, Maes P, Van Ranst M, Wollants E. Sequencing directly from antigen-detection rapid diagnostic tests in Belgium, 2022: a gamechanger in genomic surveillance? Euro Surveill. 2023;28:91. DOIPubMedGoogle Scholar
- Paull JS, Petros BA, Brock-Fisher TM, Jalbert SA, Selser VM, Messer KS, et al. Optimisation and evaluation of viral genomic sequencing of SARS-CoV-2 rapid diagnostic tests: a laboratory and cohort-based study. Lancet Microbe. 2024;5:e468–77. DOIPubMedGoogle Scholar
- Nguyen PV, Carmola LR, Wang E, Bassit L, Rao A, Greenleaf M, et al. SARS-CoV-2 molecular testing and whole genome sequencing following RNA recovery from used BinaxNOW COVID-19 antigen self tests. J Clin Virol. 2023;162:
105426 . DOIPubMedGoogle Scholar - Macori G, Russell T, Barry G, McCarthy SC, Koolman L, Wall P, et al. Inactivation and recovery of high quality RNA from positive SARS-CoV-2 rapid antigen tests suitable for whole virus genome sequencing. Front Public Health. 2022;10:
863862 . DOIPubMedGoogle Scholar - Health Innovation Program. ZIP codes by rural and urban groupings: HIPxChange [cited 2025 Jan 3]. https://hipxchange.org/toolkit/ruralurbangroups
- Coelho FF, da Silva MA, Lopes TB, Polatto JM, de Castro NS, Andrade LAF, et al. SARS-CoV-2 rapid antigen test based on a new anti-nucleocapsid protein monoclonal antibody: development and real-time validation. Microorganisms. 2023;11:2422. DOIPubMedGoogle Scholar
- US Census Bureau. Geographic areas reference manual. Washington: the Bureau; 1994.
- Lu X, Wang L, Sakthivel SK, Whitaker B, Murray J, Kamili S, et al. US CDC real-time reverse transcription PCR panel for detection of severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis. 2020;26:1654–65. DOIPubMedGoogle Scholar
- Patel H, Monzón S, Varona S, Espinosa-Carrasco J, Garcia MU, Heuer ML, et al. nf-core/viralrecon: nf-core/viralrecon v2.6.0–rhodium raccoon [cited 2024 May 29]. https://zenodo.org/record/7764938
- Ewels PA, Peltzer A, Fillinger S, Patel H, Alneberg J, Wilm A, et al. The nf-core framework for community-curated bioinformatics pipelines. Nat Biotechnol. 2020;38:276–8. DOIPubMedGoogle Scholar
- Wisconsin State Laboratory of Hygiene. SARS-CoV-2 wastewater genomic dashboard [cited 2024 Jun 7]. https://dataportal.slh.wisc.edu/sc2-ww-dashboard
- Aksamentov I, Roemer C, Hodcroft E, Neher R. Nextclade: clade assignment, mutation calling and quality control for viral genomes. J Open Source Softw. 2021;6:3773. DOIGoogle Scholar
- Tosta S, Moreno K, Schuab G, Fonseca V, Segovia FMC, Kashima S, et al. Global SARS-CoV-2 genomic surveillance: What we have learned (so far). Infect Genet Evol. 2023;108:
105405 . DOIPubMedGoogle Scholar - Public Health Madison & Dane County. Respiratory illness dashboard [cited 2024 Jun 7]. https://publichealthmdc.com/health-services/respiratory-illness/dashboard
- US Postal Service. Postal Service delivery performance continues to average 2.6 days [cited 2024 Nov 13]. https://about.usps.com/newsroom/national-releases/2023/1222-usps-delivery-performance-continues-to-average-2-6-days.htm
- Chen C, Nadeau S, Yared M, Voinov P, Xie N, Roemer C, et al. CoV-Spectrum: analysis of globally shared SARS-CoV-2 data to identify and characterize new variants. Bioinformatics. 2022;38:1735–7. DOIPubMedGoogle Scholar
- Food and Drug Administration. Influenza diagnostic tests [cited 2024 Aug 2]. https://www.fda.gov/medical-devices/in-vitro-diagnostics/influenza-diagnostic-tests
- Therapeutic Goods Administration. Respiratory combo panel RSV/SARS-CoV-2/Influenza A/B Rapid Antigen Test Kit RAT-19 (self-test) (nasal swab) (combination self‐tests) [cited 2024 Nov 14]. https://www.tga.gov.au/resources/covid-19-test-kits/respiratory-combo-panel-rsv-sars-cov-2-influenza-ab-rapid-antigen-test-kit-rat-19-self-test-nasal-swab-combination-self-tests
- Therapeutic Goods Administration. COVID-19, Influenza A/B & RSV Antigen Nasal Test Kit for self-testing (Biolink Biopen) [cited 2024 Nov 14]. https://www.tga.gov.au/resources/covid-19-test-kits/covid-19-influenza-ab-rsv-antigen-nasal-test-kit-self-testing-biolink-biopen
- Smith-Jeffcoat SE, Mellis AM, Grijalva CG, Talbot HK, Schmitz J, Lutrick K, et al.; RVTN-Sentinel Study Group. SARS-CoV-2 viral shedding and rapid antigen test performance— respiratory virus transmission network, November 2022–May 2023. MMWR Morb Mortal Wkly Rep. 2024;73:365–71. DOIPubMedGoogle Scholar