Volume 31, Number 2—February 2025
Dispatch
Infection by Tickborne Bacterium Candidatus Midichloria Associated with First Trimester Pregnancy Loss, Tennessee, USA
Figure 3
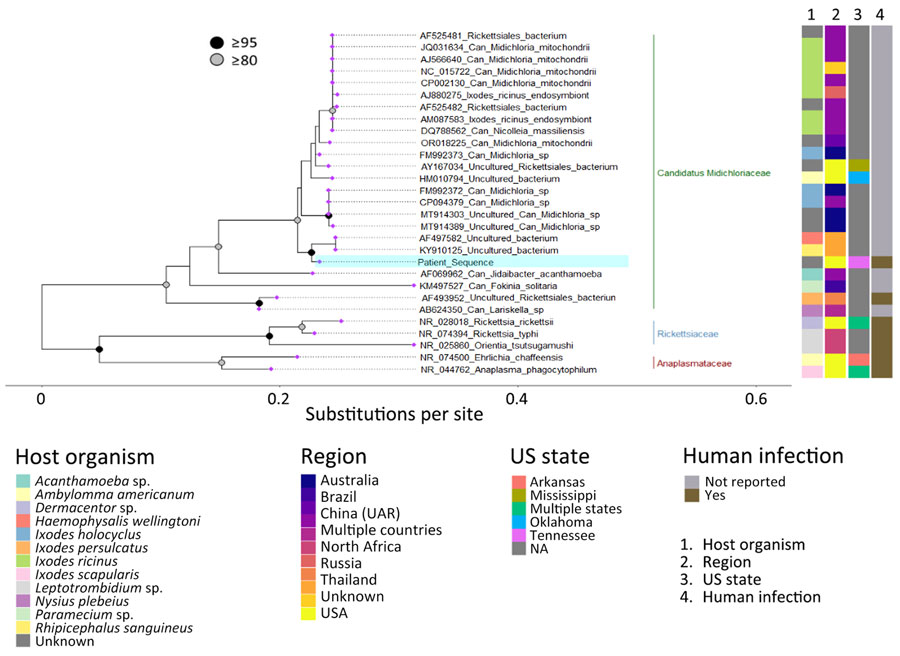
Figure 3. Phylogenetic analysis of a patient-derived sequence (highlighted in blue) in study of infection by tickborne bacterium Candidatus Midichloria associated with first trimester pregnancy loss, Tennessee, USA. Phylogenetic tree was generated using IQ-TREE (https://www.iqtree.org) with MAFFT-aligned (https://www.mafft.cbrc.jp/alignment/software) representative V1/V2 regions of 16S rRNA gene sequences from organisms within Rickettsiales and visualized with ggtree in R version 4.2.2 (The R Project for Statistical Computing, https://www.r-project.org). Sequences represented families Can. Midichloriaceae (green bar), Anaplasmataceae (red bar), and Rickettsiaceae (blue bar); most proximal sequences by BLAST analysis (https://blast.ncbi.nlm.nih.gov), including those used during clinical identification; and sequences previously associated with human specimens. GenBank accession numbers are indicated before the species name. Associated metadata indicate host tick, country reported, US state if applicable, and whether previously associated with a human infection. UAR, Uighur Autonomous Region. Nodes with >95% bootstrap support are in black, those with 80%–94.9% support are in gray. Tip labels are purple.