Simultaneous Detection of Sarcocystis hominis, S. heydorni, and S. sigmoideus in Human Intestinal Sarcocystosis, France, 2021–2024
Maxime Moniot, Patricia Combes, Damien Costa, Nicolas Argy, Marie-Fleur Durieux, Thomas Nicol, Céline Nourrisson, and Philippe Poirier

Author affiliation: CHU Clermont-Ferrand Service de Parasitologie-Mycologie, Clermont-Ferrand, France (M. Moniot); CHU Clermont-Ferrand Centre National de Référence des cryptosporidioses, microsporidies et autres protozooses digestives, Clermont-Ferrand (M. Moniot, P. Combes, D. Costa, C. Nourrisson, P. Poirier); Université Rouen Normandie, Rouen, France (D. Costa); CHU Rouen Centre National de Référence Cryptosporidioses, Microsporidies et autres protozooses digestives (Centre coordonnateur), Rouen (D. Costa); Hopital Bichat-Claude Bernard, Paris, France (N. Argy); MERIT UMR 216, IRD, Faculté de pharmacie de Paris, Université Paris Cité, Paris (N. Argy); Centre de biologie et de recherche en santé, Hôpital Universitaire Dupuytren, Limoges, France (M. Durieux); CHU Angers, Angers, France (T. Nicol); Microbes, Intestin, Inflammation et Susceptibilité de l’Hôte, UMR Inserm/Université Clermont Auvergne U1071, USC INRAE 1382, Clermont-Ferrand (C. Nourrisson, P. Poirier)
Main Article
Figure 2
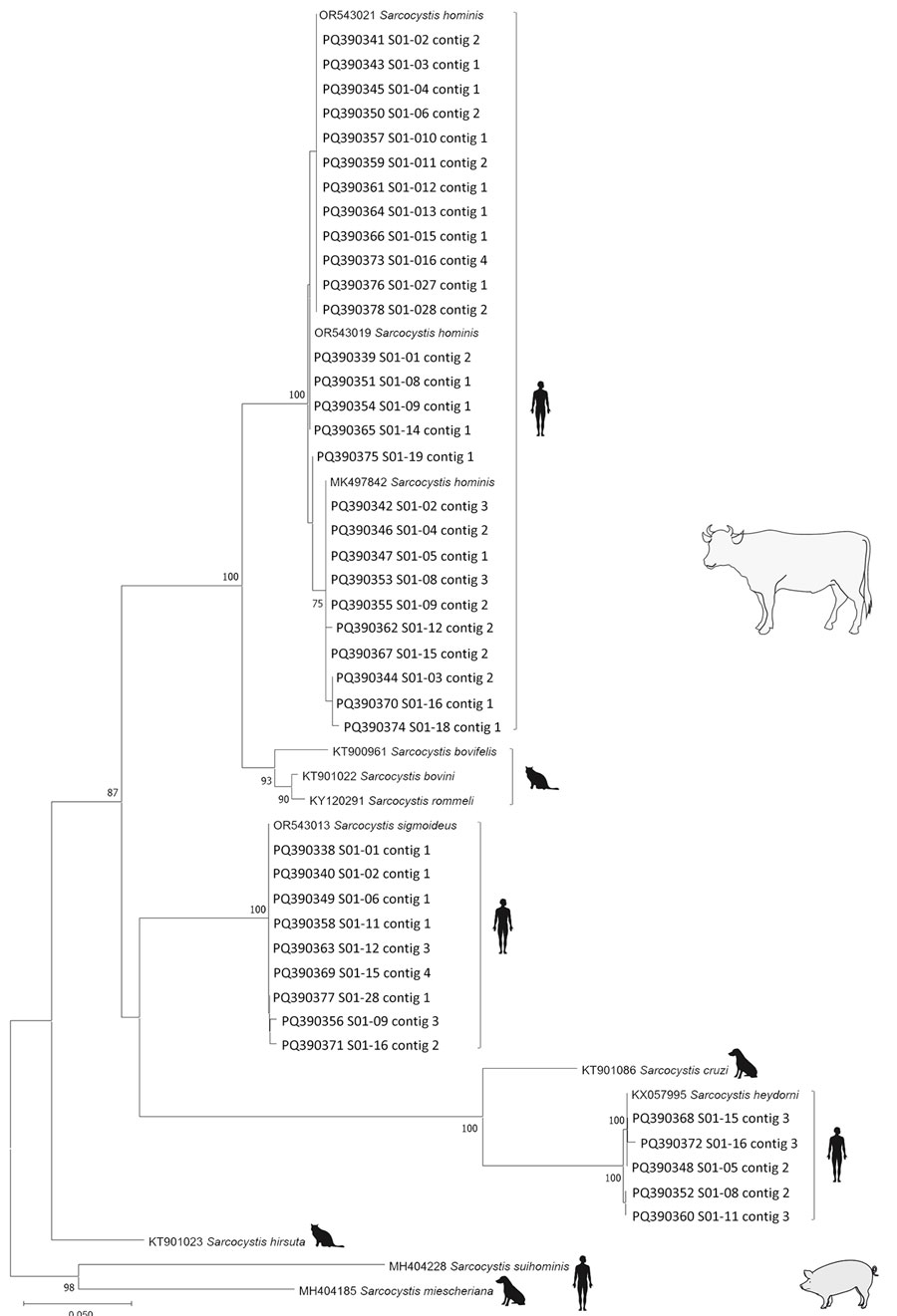
Figure 2. Phylogenetic tree for human Sarcocystis spp. from human intestinal sarcocystosis, France, 2021–2024. Tree is based on 54 partial mitochondrial cytochrome c oxidase subunit I gene sequences from patients compared with reference sequences from GenBank. The 41 sequences of 332 bp generated in this study are identified by patient numbers (i.e., S01-02); GenBank accession numbers are provided for reference sequences. This analysis included the 10 taxa described in pigs and cattle (intermediate hosts, blank illustrations) with the corresponding definitive host (humans, felids, or canids, black illustrations). The tree was inferred by using the neighbor-joining method and rooted on the species whose pigs serve as the intermediate hosts, S. miescheriana and S. suihominis. Evolutionary distances were computed using the Tamura-Nei method. Branch consensus support is expressed as percentage from 1,000 bootstraps and is reported next to the branches; branch support values <75% were not included. Scale bar indicates base substitutions per site.
Main Article
Page created: February 04, 2025
Page updated: February 28, 2025
Page reviewed: February 28, 2025
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
