Volume 31, Number 6—June 2025
Synopsis
Genomic Surveillance of Climate-Amplified Cholera Outbreak, Malawi, 2022–2023
Figure 2
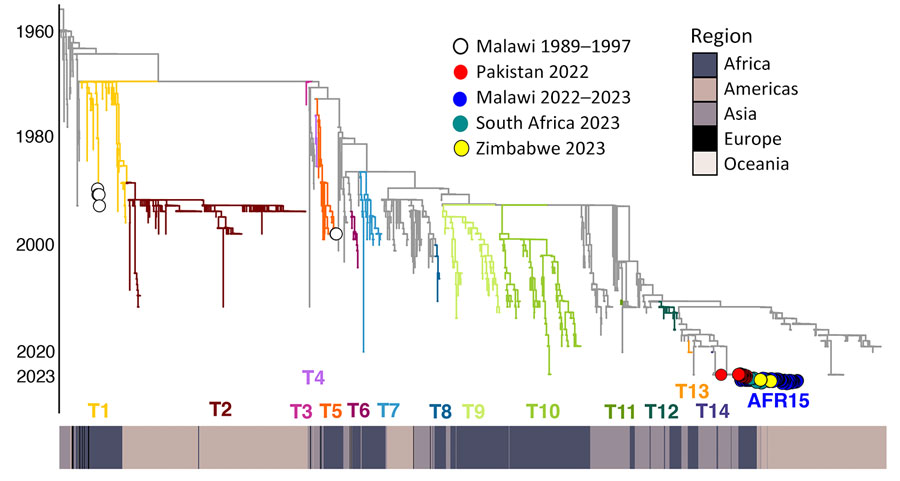
Figure 2. Phylogenetic history of cholera outbreaks within Malawi shown as part of a time-scaled maximum likelihood global phylogeny of 2,778 cholera genomes. Clade branches are colored by the previous 12 introduction events involving Africa (T1–T14 and AFR15) as described by Weill et al. (9). We denote the clade containing the genomes from the 2022–2023 outbreaks in Malawi, South Africa, and Zimbabwe as the AFR15 lineage. Heat map below the tree shows continent of sample origin.
References
- Kanungo S, Azman AS, Ramamurthy T, Deen J, Dutta S. Cholera. Lancet. 2022;399:1429–40. DOIPubMedGoogle Scholar
- World Health Organization. Outbreaks and other emergencies bulletin, week 30: 24–30 July 2023. 2023 [cited 2025 Feb 24]. https://www.afro.who.int/health-topics/disease-outbreaks/outbreaks-and-other-emergencies-updates
- World Health Organization. Disease outbreak news; cholera—global situation. 2022 Dec 16 [cited 2025 Feb 24]. https://www.who.int/emergencies/disease-outbreak-news/item/2022-DON426
- World Health Organization. Disease outbreak news; cholera—global situation. 2023 Feb 11. [cited 2025 Feb 24]. https://www.who.int/emergencies/disease-outbreak-news/item/2023-DON437
- Msyamboza KP, Kagoli M, M’bang’ombe M, Chipeta S, Masuku HD. Cholera outbreaks in Malawi in 1998‒2012: social and cultural challenges in prevention and control. J Infect Dev Ctries. 2014;8:720–6. DOIPubMedGoogle Scholar
- World Health Organization African Region. Weekly regional cholera bulletin. 2024 Apr 1 [cited 2025 Feb 24]. https://iris.who.int/bitstream/handle/10665/376526/AFRO%20Cholera%20Bulletin.58.pdf
- World Health Organization. Disease outbreak news: cholera—Malawi. 2023 Feb 9 [cited 2025 Feb 24]. https://www.who.int/emergencies/disease-outbreak-news/item/2022-DON435
- Mutreja A, Kim DW, Thomson NR, Connor TR, Lee JH, Kariuki S, et al. Evidence for several waves of global transmission in the seventh cholera pandemic. Nature. 2011;477:462–5. DOIPubMedGoogle Scholar
- Weill FX, Domman D, Njamkepo E, Tarr C, Rauzier J, Fawal N, et al. Genomic history of the seventh pandemic of cholera in Africa. Science. 2017;358:785–9. DOIPubMedGoogle Scholar
- Inzaule SC, Tessema SK, Kebede Y, Ogwell Ouma AE, Nkengasong JN. Genomic-informed pathogen surveillance in Africa: opportunities and challenges. Lancet Infect Dis. 2021;21:e281–9. DOIPubMedGoogle Scholar
- Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–90. DOIPubMedGoogle Scholar
- Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–93. DOIPubMedGoogle Scholar
- Tonkin-Hill G, Lees JA, Bentley SD, Frost SDW, Corander J. Fast hierarchical Bayesian analysis of population structure. Nucleic Acids Res. 2019;47:5539–49. DOIPubMedGoogle Scholar
- Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43:
e15 . DOIPubMedGoogle Scholar - Shen W, Le S, Li Y, Hu F. SeqKit: a cross-platform and ultrafast toolkit for FASTA/Q file manipulation. PLoS One. 2016;11:
e0163962 . DOIPubMedGoogle Scholar - Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol. 2018;35:1547–9. DOIPubMedGoogle Scholar
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–74. DOIPubMedGoogle Scholar
- Sagulenko P, Puller V, Neher RA. TreeTime: maximum-likelihood phylodynamic analysis. Virus Evol. 2018;4:
vex042 . DOIPubMedGoogle Scholar - Chaguza C, Chibwe I, Chaima D, Musicha P, Ndeketa L, Kasambara W, et al. Genomic insights into the 2022–2023 Vibrio cholerae outbreak in Malawi. Nat Commun. 2024;15:6291.
- Smith AM, Sekwadi P, Erasmus LK, Lee CC, Stroika SG, Ndzabandzaba S, et al. Imported cholera cases, South Africa, 2023. Emerg Infect Dis. 2023;29:1687–90. DOIPubMedGoogle Scholar
- Olatunji G, Kokori E, Moradeyo A, Olatunji D, Ajibola F, Otolorin O, et al. A perspective on the 2023 cholera outbreaks in Zimbabwe: implications, response strategies, and policy recommendations. J Epidemiol Glob Health. 2024;14:243–8. DOIPubMedGoogle Scholar
- Rambaut A, Lam TT, Max Carvalho L, Pybus OG. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen). Virus Evol. 2016;2:
vew007 . DOIPubMedGoogle Scholar - Gill MS, Lemey P, Faria NR, Rambaut A, Shapiro B, Suchard MA. Improving Bayesian population dynamics inference: a coalescent-based model for multiple loci. Mol Biol Evol. 2013;30:713–24. DOIPubMedGoogle Scholar
- Suchard MA, Lemey P, Baele G, Ayres DL, Drummond AJ, Rambaut A. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 2018;4:
vey016 . DOIPubMedGoogle Scholar - Baele G, Li WL, Drummond AJ, Suchard MA, Lemey P. Accurate model selection of relaxed molecular clocks in bayesian phylogenetics. Mol Biol Evol. 2013;30:239–43. DOIPubMedGoogle Scholar
- Mavian C, Paisie TK, Alam MT, Browne C, Beau De Rochars VM, Nembrini S, et al. Toxigenic Vibrio cholerae evolution and establishment of reservoirsin aquatic ecosystems. Proceedings of the National Academy of Sciences. 2020:201918763.
- Mavian CN, Tagliamonte MS, Alam MT, Sakib SN, Cash MN, Moir M, et al. Ancestral origin and dissemination dynamic of reemerging toxigenic Vibrio cholerae, Haiti. Emerg Infect Dis. 2023;29:2072–82. DOIPubMedGoogle Scholar
- Yu GC, Smith DK, Zhu HC, Guan Y, Lam TTY. GGTREE: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol. 2017;8:28–36. DOIGoogle Scholar
- Bielejec F, Rambaut A, Suchard MA, Lemey P. SPREAD: spatial phylogenetic reconstruction of evolutionary dynamics. Bioinformatics. 2011;27:2910–2. DOIPubMedGoogle Scholar
- Bhatt S, Ferguson N, Flaxman S, Gandy A, Mishra S, Scott JA. Semi-mechanistic Bayesian modelling of COVID-19 with renewal processes. J R Stat Soc Ser A Stat Soc. 2023;186:601–15. DOIGoogle Scholar
- Cori A, Ferguson NM, Fraser C, Cauchemez S. A new framework and software to estimate time-varying reproduction numbers during epidemics. Am J Epidemiol. 2013;178:1505–12. DOIPubMedGoogle Scholar
- Cartalis C, Feidas H, Glezakou M, Proedrou M, Chrysoulakis N. Use of earth observation in support of environmental impact assessments: prospects and trends. Environ Sci Policy. 2000;3:287–94.
- Tarpanelli A, Mondini AC, Camici S. Effectiveness of Sentinel-1 and Sentinel-2 for flood detection assessment in Europe. Nat Hazards Earth Syst Sci. 2022;22:2473–89. DOIGoogle Scholar
- Li S, Sun D, Goldberg MD, Sjoberg B, Santek D, Hoffman JP, et al. Automatic near real-time flood detection using Suomi-NPP/VIIRS data. Remote Sens Environ. 2018;204:672–89.
- Turner B. International Air Transport Association (IATA). In: Turner B, editor. The statesman’s yearbook 2008: the politics, cultures and economies of the world. London: Palgrave Macmillan UK; 2007. p. 48.
- Sim EM, Martinez E, Blackwell GA, Pham D, Millan G, Graham RMA, et al. Genomes of Vibrio cholerae O1 serotype Ogawa associated with current cholera activity in Pakistan. Microbiol Resour Announc. 2023;12:
e0088722 . DOIPubMedGoogle Scholar - World Health Organization Eastern Mediterranean Region. Epidemic and pandemic-prone diseases: acute watery diarrhoea/cholera updates (16–31 March 2023). 2023 [cited 2025 Feb 24]. https://www.emro.who.int/pandemic-epidemic-diseases/cholera/acute-watery-diarrhoeacholera-updates-1631-march-2023.html
- Jamil H, Liaqat A, Lareeb I, Tariq W, Jaykumar V, Kumar L, et al. Monsoon and cholera outbreaks in Pakistan: a public health concern during a climate catastrophe. IJS Global Health. 2023;6:e105.
- Global Task Force on Cholera Control. Ending cholera: a global roadmap to 2030. 2017 [cited https://www.gtfcc.org/wp-content/uploads/2019/10/gtfcc-ending-cholera-a-global-roadmap-to-2030.pdf
1These authors contributed equally to this article.
Page created: May 01, 2025
Page updated: May 27, 2025
Page reviewed: May 27, 2025
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.