Persistence of SARS-CoV-2 Alpha Variant in White-Tailed Deer, Ohio, USA
Natalie N. Tarbuck
1, Sofya K. Garushyants
1, Dillon S. McBride, Patricia M. Dennis, John Franks, Karlie Woodard, Austin Shamblin, Michael G. Sovic, Derek T. Collins, Kyle Van Why, Richard J. Webby, Martha I. Nelson, and Andrew S. Bowman

Author affiliation: The Ohio State University, Columbus, Ohio, USA (N.N. Tarbuck, D.S. McBride, P.M. Dennis, A. Shamblin, M.G. Sovic, A.S. Bowman); National Institutes of Health, Bethesda, Maryland, USA (S.K. Garushyants, M.I. Nelson); Cleveland Metroparks Zoo, Cleveland, Ohio, USA (P.M. Dennis); St. Jude Children's Research Hospital, Memphis, Tennessee, USA (J. Franks, K. Woodard, R.J. Webby); Animal and Plant Health Inspection Service, Fort Collins, Colorado, USA (D.T. Collins); Animal and Plant Health Inspection Service, Harrisburg, Pennsylvania, USA (K. Van Why).
Main Article
Figure 2
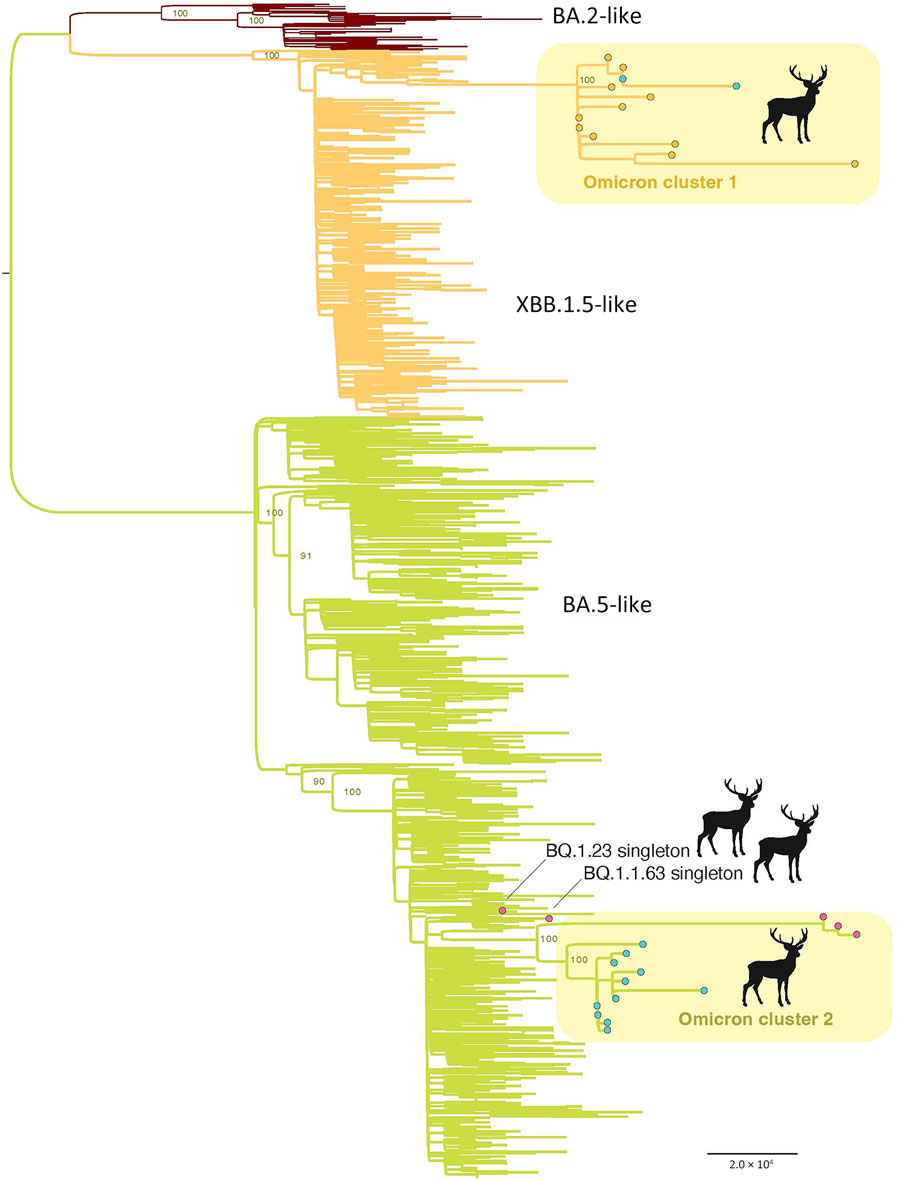
Figure 2. Maximum-likelihood tree inferred for the complete genome sequences of 31 SARS-CoV-2 viruses collected from white-tailed deer and a random subsample of 1,000 SARS-CoV-2 viruses collected from humans from northeast Ohio, November 1, 2022–March 30, 2023. Branches are shaded according to 3 categories of Omicron viruses: red, BA.2-like; orange, XBB.1.5-like; and green, BA.5-like. Circles at tips indicate the deer viruses and are shaded according to region: orange, R1; pink, R5; and blue, R9. Two deer clusters (Omicron cluster 1 and 2) are labeled, along with the 2 single deer singleton viruses. All branch lengths are drawn to scale and bootstrap values are provided for key nodes. Scale bar represents substitutions per site.
Main Article
Page created: May 13, 2025
Page updated: June 25, 2025
Page reviewed: June 25, 2025
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
