Volume 31, Number 7—July 2025
Research
Persistence of SARS-CoV-2 Alpha Variant in White-Tailed Deer, Ohio, USA
Figure 3
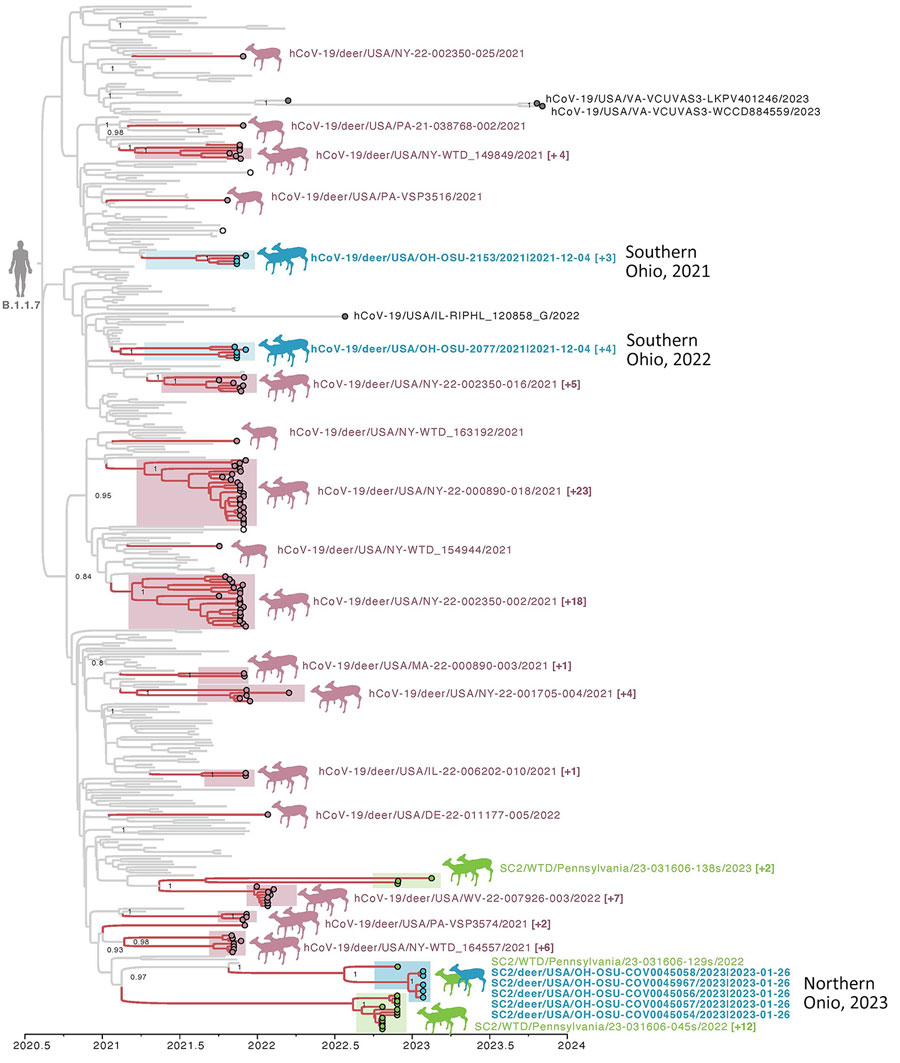
Figure 3. Time-scaled maximum clade credibility tree of the Alpha variant of SARS-CoV-2 viruses circulating in WTD in northeast Ohio, USA. The tree was inferred by using a host-specific local clock for the complete genome sequences of 362 SARS-CoV-2 clade B.1.1.7 viruses sampled December 29, 2020–November 3, 2023. Branches are shaded by host: gray, human or mink; pink, WTD. Circles at tips are shaded by location and source: blue, WTD from Ohio; green, WTD from Pennsylvania (2022–2023); pink, WTD from other states (or Pennsylvania in 2021); white, mink (Lithuania and Poland); and gray, human (2022–2023). Deer transmission clusters are shaded similarly and indicated by paired deer illustrations. Single-deer illustrations indicate singleton introductions into WTD. Singleton WTD viruses are labeled, and a representative virus is provided for each WTD transmission cluster, with the number of additional viruses in the cluster in parentheses. Posterior probabilities are provided for key nodes. WTD, white-tailed deer.
1These authors contributed equally to this article.