Volume 31, Number 7—July 2025
Research
Persistence of SARS-CoV-2 Alpha Variant in White-Tailed Deer, Ohio, USA
Figure 4
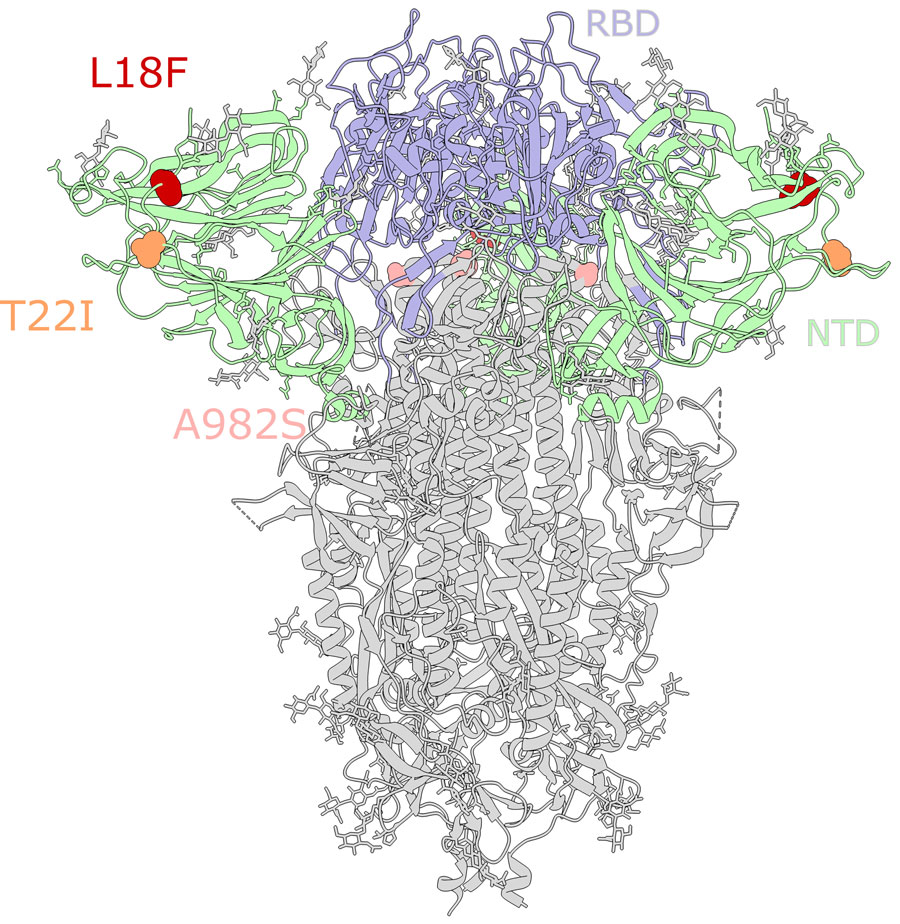
Figure 4. Mutations in the spike glycoprotein of SARS-CoV-2 viruses circulating in white-tailed deer in northeast Ohio, USA. SARS-CoV-2 spike glycoprotein structure obtained from the Protein Data Base (identification no. 7jji; https://www.rcsb.org). RBD and NTD are labeled. Circles indicate the location of mutations in the SARS-CoV-2 clade B.1.1.7 viruses collected from white-tailed deer in Ohio and other US states: red, L18F; orange, T22I; pink, A982S. Mutation S12F (located in NTD) is not shown because it is located at the flexible N terminus, for which a 3D structure is not available. NTD, N-terminal domain; RBD, receptor-binding domain.
1These authors contributed equally to this article.
Page created: May 13, 2025
Page updated: June 25, 2025
Page reviewed: June 25, 2025
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.