Volume 31, Number 8—August 2025
Research Letter
Nipah Virus Antibodies in Bats, the Philippines, 2013–2022
Abstract
In 2014, an outbreak of zoonotic Nipah virus (NiV) occurred on Mindanao Island, the Philippines. We investigated the prevalence of NiV in Philippine bats. Because neutralizing antibodies were detected in insectivorous bats on Siargao Island, public health officials should consider that the distribution range of NiV is not limited to Mindanao Island.
Nipah virus (NiV; family Paramyxoviridae, genus Henipavirus) was first discovered in 1998–1999. Officials in Malaysia and Singapore identified it as a causative virus of severe respiratory disease in pigs and highly fatal encephalitis or respiratory disease in humans (1). Subsequently, Bangladesh and India have reported sporadic outbreaks of the virus almost annually (2,3). Direct bat-to-human transmission is assumed in those outbreaks; however, human-to-human transmission through concentrated contact has also been reported (3).
In Southeast Asia, some frugivorous bat species (mainly of the genus Pteropus) and several insectivorous bat species (genera Hipposideros, Scotophilus, and Rhinolophus) are reservoirs of the virus, which has led to its widespread transmission (4–6). In 2014, in Sultan Kudarat Province, which is located in the southern part of Mindanao Island in the Philippines, 10 horses died, and serious infections occurred in 17 humans, mainly in those who had slaughtered horses or consumed horse meat (7). The humans who died had acute encephalitis syndrome, a severe influenza-like illness, or meningitis, and the etiology was diagnosed as henipavirus infection on the basis of neutralizing antibody detection in patient serum samples. One patient had a short 71-bp fragment sequence that was 99% homologous to the NiV strain from Malaysia, suggesting that NiV was the etiologic virus (7). The likely source of infection in horses is bats, which are a natural host of the virus.
Residual serum samples used in epidemiologic studies of bat-derived viruses conducted before 2019 were reused in this NiV epidemiologic study (8). In addition, we conducted new bat trapping at the end of 2022. In each study, we collected specimens from wild bats.
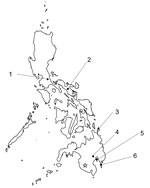
We attempted to detect NiV-neutralizing antibodies by using serum samples collected from bats in 6 regions of the Philippines, spanning from north to south (Figure). We determined the neutralization titer of each serum sample by using a surrogate assay without an infectious NiV, as previously established (9). Using vesicular stomatitis virus expressing secreted alkaline phosphatase pseudotyped with G and F proteins of the NiV strain from Malaysia (VSV-NiV-SEAP) (9), we determined the titer of the neutralizing antibody. Moreover, we performed detection of NiV RNA with reverse transcription PCR by using consensus primers that widely detect paramyxoviruses (PAR-F1, PAR-F2, and PAR-R) (Appendix) (10).
In total, we diluted 326 bat serum samples 80-fold and screened for VSV-NiV-SEAP (Table) (9). We subjected 4 serum samples that tested reactive in screening to serial dilution. We determined antibody titers as values of 16, 41, 47, and 141, which are shown as the reciprocal of the serum dilution factor at which SEAP activity was suppressed by >75% after VSV-NiV-SEAP entered the cells (9). We obtained positive samples from the insectivorous bat Hipposideros diadema, which was captured on Siargao Island (Figure). We used a similar surrogate system to detect neutralizing antibodies against Hendra virus. The same 4 serum samples showed cell entry inhibition rates ranging from 35.2% to 63.1% against VSV pseudotyped with Hendra virus G and F proteins. Those results were weaker than those obtained for VSV-NiV-SEAP in the screening (Appendix Table). However, because of an insufficient volume of serum samples, we could not perform titration by serial dilution. In contrast, we did not detect any neutralizing antibodies in bats from Mindanao Island or elsewhere (Table). Moreover, we did not detect any viral RNA in reverse transcription PCR targeting paramyxoviruses (including NiV and Hendra virus) using RNA extracted from the 252 samples (collected from serum or spleen) (Table).
In this study, we investigated the prevalence of NiV with bat serum samples collected from 6 regions in the Philippines (Figure). We did not detect any antibodies on Mindanao Island, where the henipavirus outbreak occurred, which may be partially because we could not capture and study the primary reservoir, Pteropus bats, which fly and migrate at high altitudes. However, we detected NiV antibodies in 4 samples from 1 insectivorous bat species on Siargao Island (Table), which is geographically close, indicating that the distribution range of NiV is not limited to within Mindanao Island.
Antibodies have been reported from other Hipposideros bat species closely related to H. diadema (5). We also captured a species (Scotophilus kuhlii) other than Pteropus bats, for which antibodies were similarly detected in bats in previous reports (5), but we did not detect any antibodies. In contrast, we could not detect viral RNA in all samples because of the small number of samples. We consider it crucial to obtain more viral genetic information to understand the nature of the virus responsible for the henipavirus epidemic in the Philippines and to take countermeasures. More detailed surveys with larger sample sizes on Mindanao Island and surrounding areas are needed. Surveillance of NiV carriage in bats in the Philippines is necessary to characterize the virus, investigate risk factors for future outbreaks of henipavirus, and implement control measures.
Dr. Kaku is a researcher at the National Institute of Infectious Diseases of Japan. His research interests include epidemiologic studies of henipavirus and rabies virus, analysis of viral pathogenicity mechanisms, and development of diagnostic systems.
Acknowledgments
We thank Momoko Ogata for assistance in this study. We thank Eduardo Eres, James D.V. Alvarez, Yuki Sugiura, and Roberto Puentespina Jr. for assistance in collecting samples. We also thank Editage (https://www.editage.com) for the English language editing.
Wild bats were captured under a permit issued by the Department of Environment and Natural Resources to the University of the Philippines Los Baños for this research purpose (Wildlife Gratuitous permit nos. RXI-2013-06, R13-2019-27, and R5-2019-105). Furthermore, for every scientific expedition undertaken by the authors to capture bats, a permit was issued by the Biodiversity Management Bureau. Each scientific expedition to capture bats was also covered by a permit granted by the local regional office of the Department of Environment and Natural Resources. The procedures for serum and spleen sample collection after euthanasia of the captured bats were carried out based on the guidance of the institutional animal care and use committee of the University of the Philippines Los Baños.
This study was supported by grants from the Takeda Science Foundation, Kanae Foundation for the Promotion of Medical Science, Japan Society for the Promotion of Science (JSPS KAKENHI, grant no. JP22K06016 and 19KK0242), Health Labour Sciences Research (grant no. 23HA2004), and the Japan Science and Technology Agency (JST SICORP e-ASIA, grant no. JPMJSC20U2).
References
- Ang BSP, Lim TCC, Wang L. Nipah virus infection. J Clin Microbiol. 2018;56:e01875–17. DOIPubMedGoogle Scholar
- As AK, Sahay RR, Radhakrishnan C, P S, Kandath S, Patil DY, et al. Clinico-epidemiological presentations and management of Nipah virus infection during the outbreak in Kozhikode district, Kerala state, India 2023. J Med Virol. 2024;96:
e29559 . DOIPubMedGoogle Scholar - Gurley ES, Montgomery JM, Hossain MJ, Bell M, Azad AK, Islam MR, et al. Person-to-person transmission of Nipah virus in a Bangladeshi community. Emerg Infect Dis. 2007;13:1031–7. DOIPubMedGoogle Scholar
- Gokhale MD, Sreelekshmy M, Sudeep AB, Shete A, Jain R, Yadav PD, et al. Detection of possible Nipah virus infection in Rousettus leschenaultii and Pipistrellus Pipistrellus bats in Maharashtra, India. J Infect Public Health. 2021;14:1010–2. DOIPubMedGoogle Scholar
- Plowright RK, Becker DJ, Crowley DE, Washburne AD, Huang T, Nameer PO, et al. Prioritizing surveillance of Nipah virus in India. PLoS Negl Trop Dis. 2019;13:
e0007393 . DOIPubMedGoogle Scholar - Reynes JM, Counor D, Ong S, Faure C, Seng V, Molia S, et al. Nipah virus in Lyle’s flying foxes, Cambodia. Emerg Infect Dis. 2005;11:1042–7. DOIPubMedGoogle Scholar
- Ching PK, de los Reyes VC, Sucaldito MN, Tayag E, Columna-Vingno AB, Malbas FF Jr, et al. Outbreak of henipavirus infection, Philippines, 2014. Emerg Infect Dis. 2015;21:328–31. DOIPubMedGoogle Scholar
- Taniguchi S, Maeda K, Horimoto T, Masangkay JS, Puentespina R Jr, Alvarez J, et al. First isolation and characterization of pteropine orthoreoviruses in fruit bats in the Philippines. Arch Virol. 2017;162:1529–39. DOIPubMedGoogle Scholar
- Kaku Y, Noguchi A, Marsh GA, Barr JA, Okutani A, Hotta K, et al. Second generation of pseudotype-based serum neutralization assay for Nipah virus antibodies: sensitive and high-throughput analysis utilizing secreted alkaline phosphatase. J Virol Methods. 2012;179:226–32. DOIPubMedGoogle Scholar
- Tong S, Chern SW, Li Y, Pallansch MA, Anderson LJ. Sensitive and broadly reactive reverse transcription-PCR assays to detect novel paramyxoviruses. J Clin Microbiol. 2008;46:2652–8. DOIPubMedGoogle Scholar
Figure
Table
Cite This ArticleOriginal Publication Date: July 16, 2025
Table of Contents – Volume 31, Number 8—August 2025
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Shumpei Watanabe, Department of Microbiology, Faculty of Veterinary Medicine, Okayama University of Science, 1-3 Ikoinooka, Imabari, Ehime 794-8555, Japan
Top